Abstract
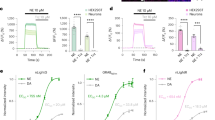
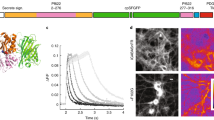
Single-wavelength fluorescent reporters allow visualization of specific neurotransmitters with high spatial and temporal resolution. We report variants of intensity-based glutamate-sensing fluorescent reporter (iGluSnFR) that are functionally brighter; detect submicromolar to millimolar amounts of glutamate; and have blue, cyan, green, or yellow emission profiles. These variants could be imaged in vivo in cases where original iGluSnFR was too dim, resolved glutamate transients in dendritic spines and axonal boutons, and allowed imaging at kilohertz rates.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
All data from this study are available upon request. All constructs have been deposited at Addgene (#106174–106206; hSynapsin1, FLEX-hSynapsin1, FLEX-CAG, GFAP promoters; some fusions with the red fluorescent protein mRuby3 are available). Sequences have been deposited in GenBank (MH392460, MH392461, MH392462, MH392463, MH392464, and MH392465). AAV is available from Addgene.
Change history
28 February 2019
The version of this paper originally published cited a preprint version of ref. 12 instead of the published version (Proc. Natl. Acad. Sci. USA 115, 5594–5599; 2018), which was available before this Nature Methods paper went to press. The reference information has been updated in the PDF and HTML versions of the article.
02 January 2019
In the version of this paper originally published, important figure labels in Fig. 3d were not visible. An image layer present in the authors’ original figure that included two small dashed outlines and text labels indicating ROI 1 and ROI 2, as well as a scale bar and the name of the cell label, was erroneously altered during image processing. The figure has been corrected in the HTML and PDF versions of the paper.
References
Marvin, J. S. et al. Nat. Methods 10, 162–170 (2013).
Park, S. J. H., Kim, I.-J., Looger, L. L., Demb, J. B. & Borghuis, B. G. J. Neurosci. 34, 3976–3981 (2014).
Brunert, D., Tsuno, Y., Rothermel, M., Shipley, M. T. & Wachowiak, M. J. Neurosci. 36, 6820–6835 (2016).
O’Herron, P. et al. Nature 534, 378–382 (2016).
Xie, Y. et al. J. Neurosci. 36, 1261–1272 (2016).
Bao, H. et al. Nat. Struct. Mol. Biol. 23, 67–73 (2016).
Rosa, J. M. et al. eLife 4, 728 (2015).
Enger, R. et al. Cereb. Cortex 25, 4469–4476 (2015).
Jiang, R., Diaz-Castro, B., Looger, L. L. & Khakh, B. S. J. Neurosci. 36, 3453–3470 (2016).
Pédelacq, J.-D., Cabantous, S., Tran, T., Terwilliger, T. C. & Waldo, G. S. Nat. Biotechnol. 24, 79–88 (2006).
Marvin, J. S. & Hellinga, H. W. Nat. Struct. Biol. 8, 795–798 (2001).
Helassa, N. et al. Proc. Natl Acad. Sci. USA 115, 5594–5599 (2018).
Dodge, F. A. Jr. & Rahamimoff, R. J. Physiol. (Lond.) 193, 419–432 (1967).
Abrahamsson, T., Cathala, L., Matsui, K., Shigemoto, R. & Digregorio, D. A. Neuron 73, 1159–1172 (2012).
Valera, A. M., Doussau, F., Poulain, B., Barbour, B. & Isope, P. J. Neurosci. 32, 3267–3280 (2012).
van Beugen, B. J., Gao, Z., Boele, H.-J., Hoebeek, F. & De Zeeuw, C. I. Front. Neural Circuits 7, 95 (2013).
Tang, S., Liu, J., Krasieva, T. B., Chen, Z. & Tromberg, B. J. J. Biomed. Opt. 14, 030508 (2009).
Kazemipour, A. et al. bioRxiv Preprint at https://www.biorxiv.org/content/early/2018/06/28/357269 (2018).
Borghuis, B. G. et al. J. Neurosci. 31, 2855–2867 (2011).
Pologruto, T. A., Sabatini, B. L. & Svoboda, K. Biomed. Eng. Online 2, 13 (2003).
Wilson, D. E., Whitney, D. E., Scholl, B. & Fitzpatrick, D. Nat. Neurosci. 19, 1003–1009 (2016).
Peirce, J. W. J. Neurosci. Methods 162, 8–13 (2007).
Schindelin, J. et al. Nat. Methods 9, 676–682 (2012).
Sage, D., Prodanov, D., Tinevez, J.-Y. & Schindelin, J. MIJ: making interoperability between ImageJ and Matlab possible. Poster presented at the ImageJ User Developer Conference, Luxembourg, 24–26 October 2012.
Woitecki, A. M. H. et al. J. Neurosci. 36, 2561–2570 (2016).
Silver, R. A., Cull-Candy, S. G. & Takahashi, T. J. Physiol. (Lond.) 494, 231–250 (1996).
Acknowledgements
We thank J. Macklin (Janelia Research Campus) for two-photon spectra, and K. Ritola and Janelia Virus Services for AAV production. E.C. was a Janelia Undergraduate Scholar.
Author information
Authors and Affiliations
Contributions
J.S.M., L.L.L., A.N.T., and E.C. carried out protein engineering; B.S., D.E.W., and D.F. conducted experiments on ferret visual cortex; J.A.M., S.S., and D.D. carried out neuronal culture analysis; J.P.L. assessed brightness in somatosensory cortex; K.P. and A.K. carried out high-speed imaging of the yellow variant; H.B. and E.R.C. assessed stopped-flow kinetics; N.R., F.J.U.Q., S.S.-H.W., A.W.H., and D.A.D. conducted cerebellum experiments; V.J.D. and B.G.B. conducted retina experiments; and J.S.M. and L.L.L. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
J.S.M. and L.L.L. are named on US patent 9719992, which pertains to original iGluSnFR.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14
Supplementary Video 1
Example of two-photon imaging of dendritic SFiGluSnFR.A184S activity during visual stimulation. Video is shown at 4× normal speed.
Supplementary Video 2
High-speed two-photon imaging of SF-Venus-iGluSnFR.A184V using 1,030-nm fiber laser excitation. Neurons in culture were imaged by scanned line angular projection microscopy at a 1,016-Hz frame rate. Glutamate was uncaged at two locations, 10 ms apart, starting at t = 400 ms. Saturation denotes ΔF/F0. Blue-tinted regions denote areas where excitation was blocked during high-speed imaging. Recording is a single trial, without averaging. Representative example of 4 trials.
Supplementary Video 3
Example two-photon recording of dendrites in mouse visual cortex labeled with SF-Venus-iGluSnFR.A184S, at a frame rate of 3.41 Hz. Drifting grating visual stimuli of 8 different directions, lasting 2.3 s each, were presented at 5-s intervals. Field-of-view width: 90 μm.
Rights and permissions
About this article
Cite this article
Marvin, J.S., Scholl, B., Wilson, D.E. et al. Stability, affinity, and chromatic variants of the glutamate sensor iGluSnFR. Nat Methods 15, 936–939 (2018). https://doi.org/10.1038/s41592-018-0171-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-018-0171-3
This article is cited by
-
Hydroxynorketamine, but not ketamine, acts via α7 nicotinic acetylcholine receptor to control presynaptic function and gene expression
Translational Psychiatry (2024)
-
Avoiding bias in fluorescence sensor readout
Nature Reviews Neuroscience (2024)
-
The Alzheimer’s disease risk gene BIN1 regulates activity-dependent gene expression in human-induced glutamatergic neurons
Molecular Psychiatry (2024)
-
Electrochemical and biosensor techniques to monitor neurotransmitter changes with depression
Analytical and Bioanalytical Chemistry (2024)
-
Asymmetric dysregulation of glutamate dynamics across the synaptic cleft in a mouse model of Alzheimer’s disease
Acta Neuropathologica Communications (2023)