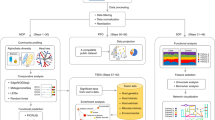
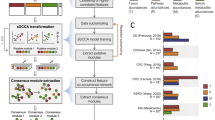
Abstract
Multi-omic insights into microbiome function and composition typically advance one study at a time. However, in order for relationships across studies to be fully understood, data must be aggregated into meta-analyses. This makes it possible to generate new hypotheses by finding features that are reproducible across biospecimens and data layers. Qiita dramatically accelerates such integration tasks in a web-based microbiome-comparison platform, which we demonstrate with Human Microbiome Project and Integrative Human Microbiome Project (iHMP) data.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Data availability
All data used are available via Qiita and EBI (where applicable). The Human Microbiome Project (HMP) and iHMP data are available via the HMP Data Analysis and Coordination Center (DACC) at https://portal.hmpdacc.org/. Analytical steps for this paper can be found at https://github.com/knightlab-analyses/qiita-paper. Additionally, the Qiita analysis can be found at https://qiita.ucsd.edu/analysis/description/15093/; note that the user must log in to Qiita to access this analysis. Source data for Supplementary Fig. 1 are available online.
References
Caporaso, J. G. et al. ISME J. 6, 1621–1624 (2012).
Thompson, L. R. et al. Nature 551, 457–463 (2017).
Halfvarson, J. et al. Nat. Microbiol. 2, 17004 (2017).
Lozupone, C. A. & Knight, R. Proc. Natl Acad. Sci. USA 104, 11436–11440 (2007).
Ley, R. E., Lozupone, C. A., Hamady, M., Knight, R. & Gordon, J. I. Nat. Rev. Microbiol. 6, 776–788 (2008).
Adams, R. I., Bateman, A. C., Bik, H. M. & Meadow, J. F. Microbiome 3, 49 (2015).
Debelius, J. et al. Genome. Biol. 17, 217 (2016).
Lozupone, C. A. et al. Genome Res. 23, 1704–1714 (2013).
Caporaso, J. G. et al. Nat. Methods 7, 335–336 (2010).
Wang, M. et al. Nat. Biotechnol. 34, 828–837 (2016).
Langille, M. G. I., Ravel, J. & Fricke, W. F. Microbiome 6, 8 (2018).
Yilmaz, P. et al. Nat. Biotechnol. 29, 415–420 (2011).
Sinha, R. et al. Nat. Biotechnol. 35, 1077–1086 (2017).
Gevers, D. et al. Cell. Host. Microbe. 15, 382–392 (2014).
Human Microbiome Project Consortium. Nature 486, 207–214 (2012).
Weingarden, A. et al. Microbiome 3, 10 (2015).
Lozupone, C. & Knight, R. Appl. Environ. Microbiol. 71, 8228–8235 (2005).
Bokulich, N. A. et al. Nat. Methods 10, 57–59 (2013).
Navas-Molina, J. A. et al. Methods Enzymol. 531, 371–444 (2013).
Amir, A. et al. mSystems 2, e00191-16 (2017).
Vázquez-Baeza, Y., Pirrung, M., Gonzalez, A. & Knight, R. Gigascience 2, 16 (2013).
Acknowledgements
We are grateful to J. Debelius, J. Jansson, D. Bazaldua, and J. Kuczynski for their help in improving Qiita via suggestion, code changes, and contributed datasets, or during the preparation of this manuscript; and to J. Gordon and his laboratory for helpful discussions. This work was supported in part by the Alfred P. Sloan Foundation (2017-9838 and 2015-13933 (R.K.)), the NIH/NIDDK (P01DK078669 (R.K.)), the NSF (DBI-1565057 and 1565100 (J.G.C. and R.K.)), the Office of Naval Research (ONR; N00014-15-1-2809 (R.K.)), and the US Army (CDMRP W81XWH-15-1-0653 (R.K.)).
Author information
Authors and Affiliations
Contributions
A.G., J.A.N.-M., T.K., D.M., Y.V.-B., G.A., J.D., S.J., A.D.S., S.B.O., J.G.S., J.S., H.H., S.P., A.R.-P., C.J.B., M.W., J.R.R., E.B., M.D., J.G.C., P.C.D., and R.K. implemented the Qiita main or the Qiita plugins code. A.G., J.A.N.-M., and Y.V.-B. conducted the example meta-analysis. All authors wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Data loaded in Qiita and uploaded to EBI.
A. Monthly studies and sample depositions to EBI-ENA via Qiita. B. Geographical distribution of the samples present in Qiita
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1, Supplementary Tables 1 and 2
Supplementary Software
SupplementarySoftware.zip contains two zip files: (1) qiita-master.zip, which is the main code for the Qiita software at the time of publication (latest version: https://github.com/biocore/qiita), and (2) qiita-paper-master.zip, which includes all steps and necessary files to reproduce all panels in Fig. 1 (live repository: https://github.com/knightlab-analyses/qiita-paper).
Source data
Rights and permissions
About this article
Cite this article
Gonzalez, A., Navas-Molina, J.A., Kosciolek, T. et al. Qiita: rapid, web-enabled microbiome meta-analysis. Nat Methods 15, 796–798 (2018). https://doi.org/10.1038/s41592-018-0141-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-018-0141-9
This article is cited by
-
Dermal injury drives a skin to gut axis that disrupts the intestinal microbiome and intestinal immune homeostasis in mice
Nature Communications (2024)
-
Bile salt hydrolase acyltransferase activity expands bile acid diversity
Nature (2024)
-
Robustness of cancer microbiome signals over a broad range of methodological variation
Oncogene (2024)
-
A genus in the bacterial phylum Aquificota appears to be endemic to Aotearoa-New Zealand
Nature Communications (2024)
-
The role of macrophyte-associated microbiomes in lacustrine wetlands: an example of the littoral zone of lake Atitlan, Guatemala
Hydrobiologia (2024)