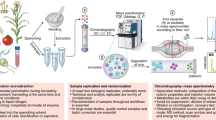
Abstract
We report XCMS-MRM and METLIN-MRM (http://xcmsonline-mrm.scripps.edu/ and http://metlin.scripps.edu/), a cloud-based data-analysis platform and a public multiple-reaction monitoring (MRM) transition repository for small-molecule quantitative tandem mass spectrometry. This platform provides MRM transitions for more than 15,500 molecules and facilitates data sharing across different instruments and laboratories.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
XCMS-MRM and METLIN-MRM are publicly available at http://xcmsonline-mrm.scripps.edu/ and http://metlin.scripps.edu/, respectively. Data and METLIN-MRM/XCMS-MRM code are available from the corresponding authors upon request.
References
Glish, G. L. & Vachet, R. W. Nat. Rev. Drug. Discov. 2, 140–150 (2003).
Patti, G. J., Yanes, O. & Siuzdak, G. Nat. Rev. Mol. Cell Biol. 13, 263–269 (2012).
Nordström, A. & Lewensohn, R. J. Neuroimmune. Pharmacol. 5, 4–17 (2010).
Strathmann, F. G. & Hoofnagle, A. N. Am. J. Clin. Pathol. 136, 609–616 (2011).
Vogeser, M. & Seger, C. Trends. Analyt. Chem. 84, 1–4 (2016).
Hoffmann, W. D. & Jackson, G. P. Annu. Rev. Anal. Chem. (Palo Alto, Calif.) 8, 419–440 (2015).
Sauer, S. & Kliem, M. Nat. Rev. Microbiol. 8, 74–82 (2010).
Nicholson, J. K., Connelly, J., Lindon, J. C. & Holmes, E. Nat. Rev. Drug. Discov. 1, 153–161 (2002).
Lange, V., Picotti, P., Domon, B. & Aebersold, R. Mol. Syst. Biol. 4, 222 (2008).
Kusebauch, U. et al. Cell 166, 766–778 (2016).
Matsumoto, M. et al. Nat. Methods 14, 251–258 (2017).
MacLean, B. et al. Bioinformatics 26, 966–968 (2010).
Cai, Y. & Weng, K. Metabolomics. 11, 1575–1586 (2015).
Tsugawa, H., Kanazawa, M., Ogiwara, A. & Arita, M. Bioinformatics 30, 2379–2380 (2014).
Guijas, C. et al. Anal. Chem. 90, 3156–3164 (2018).
Melnik, A. V. et al. Anal. Chem. 89, 7549–7559 (2017).
Domingo-Almenara, X., Montenegro-Burke, J. R., Benton, H. P. & Siuzdak, G. Anal. Chem. 90, 480–489 (2018).
Tomoiagă, B., Chindriş, M., Sumper, A., Sudria-Andreu, A. & Villafafila-Robles, R. Energies 6, 1439–1455 (2013).
Domingo-Almenara, X. et al. J. Chromatogr. A. 1409, 226–233 (2015).
De Juan, A., Vander Heyden, Y., Tauler, R. & Massart, D. L. Anal. Chim. Acta 346, 307–318 (1997).
Kessner, D., Chambers, M., Burke, R., Agus, D. & Mallick, P. Bioinformatics 24, 2534–2536 (2008).
Caroline, H. J. et al. Cell Metab. 21, 891–897 (2015).
Acknowledgements
This research was partially funded by Ecosystems and Networks Integrated with Genes and Molecular Assemblies (ENIGMA), a Scientific Focus Area Program at Lawrence Berkeley National Laboratory for the US Department of Energy, Office of Science, Office of Biological and Environmental Research under contract DE-AC02-05CH11231 (G.S.); and National Institutes of Health grants R01 GM114368-03, P30 MH062261-17 and P01 DA026146-02 (G.S.). The MRC-NIHR National Phenome Centre (NPC) is supported by the UK Medical Research Council and the National Institute for Health Research (NIHR) (England) grant MC_PC_12025. Infrastructure support for the Clinical Phenotyping Centre (CPC) is also supported by the NIHR Imperial Biomedical Research Centre. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the MRC (M.G.R., L.W., M.R.L. and J.K.N.).
Author information
Authors and Affiliations
Contributions
G.S. led the project. X.D.-A. designed and programmed the algorithms for XCMS-MRM and the statistical ranking of METLIN-MRM transitions, to which J.R.M.-B. and H.P.B. made critical contributions. J.R.M.-B. designed the analytical assays for the validation of XCMS-MRM and METLIN MRM. J.I. and A.T. acquired the experimental transitions in METLIN-MRM, and J.S. and T.T conducted the analytical assays. J.R.M.-B., C.G., L.H., J.S., A.N., M.G.-R. and L.W. analyzed the different samples in their respective laboratories. H.P.B. coordinated the online implementation of XCMS-MRM and METLIN-MRM, with assistance from A.E.A. and D.R., who programmed the online interface. X.D.-A. and J.R.M.-B. wrote the manuscript, to which C.G., J.I., A.T. and G.S. contributed. M.R.L. and J.K.N. provided advice and article revisions. All authors approved the final version of the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 METLIN-MRM screenshot.
Transitions for Tryptophan are shown. Each transition type (EO, CO or PR) is displayed in a separated table.
Supplementary Figure 2 XCMS-MRM transition-peak-processing algorithm.
a, qualitative and quantitative signals as seen in a typical MRM experiment. The quantitative transition has three peaks, but only one corresponds to the target molecule as observed from the qualitative transition signal. XCMS-MRM first examines the signal of all the transitions, including qualitative transitions, and uses that information to b, drive the integration of the qualitative transition in a more efficient manner. XCMS-MRM also subtracts the baseline that might be affecting the chromatographic signals.
Supplementary Figure 3 XCMS-MRM workflow and output.
Different type of samples including quality controls, calibration and blanks can be uploaded into XCMS-MRM for its further processing. This processing implies automated transition peak area integration, absolute concentration computation with calibration curves and results display (a, b, c). Results by XCMS-MRM (d) include analytical quality descriptors to assess the method performance and quality via quality control coefficients of variation, specificity, linear ranges and limits of detection and quantification among others. These descriptors allow validating the experiment assay to allow further cooperative biological interpretation via the cloud with the statistical results provided by XCMS-MRM. In a, the use of non-selective transition fragments (m/z 86.1) masked both leucine and isoleucine hampering its correct quantification. The use of computationally optimized selective transitions from METLIN-MRM (m/z 43.1 and 69.1) allowed their quantification in this particular example. In b, XCMS-MRM uses calibration (cal) samples to convert relative values into absolute concentration with calibration curves, allowing different calibration methods to be used. In c, XCMS-MRM integrates and aligns the detected fragment peaks of corresponding transitions. In this example, N1,N12-Diacetylspermine (DAS) was quantified in serum samples from mice before and after its administration, where as expected, DAS is seen increased in mice serum after its administration (see Methods for details).
Supplementary Figure 4 METLIN-MRM statistical ranking.
a, the transitions of the target molecule are compared to putative interfering molecules, which consist of all the molecules in the METLIN library with precursors within ±0.7 Da window the precursor of the target molecule. Selective transitions are selected based on their fragment specificity (how unique is that fragment for that molecule). This specificity or selectivity is determined with a density estimation b, where the density of the intensity of a given fragment is computed. Fragments with a low-density area indicate that this fragment has a low intensity or is not present in the interfering molecules (e.g., fragment A), and thus this fragment has a high degree of selectivity. Moreover, in cases where the most selective transition for a given molecule is not selective enough (fragment A in c) METLIN-MRM is designed to provide a second transition that provides more selectivity, in cases where the first transition is present (Fragment B in c). This approach decreases the likelihood of the two transitions appearing together if the target molecule is not present, or enables the detection of interferences if the two transitions do not show a linear relation.
Supplementary Figure 5 Results of metabolite quantification by XCMS-MRM.
Results of quantification by XCMS-MRM compared to these obtained in a certified laboratory. For each molecule, the plot shows the number of samples for each relative error range.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Tables 1–4
Rights and permissions
About this article
Cite this article
Domingo-Almenara, X., Montenegro-Burke, J.R., Ivanisevic, J. et al. XCMS-MRM and METLIN-MRM: a cloud library and public resource for targeted analysis of small molecules. Nat Methods 15, 681–684 (2018). https://doi.org/10.1038/s41592-018-0110-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-018-0110-3
This article is cited by
-
Exploring the plant lipidome: techniques, challenges, and prospects
Advanced Biotechnology (2024)
-
Guiding the choice of informatics software and tools for lipidomics research applications
Nature Methods (2023)
-
Fungal secondary metabolites in food and pharmaceuticals in the era of multi-omics
Applied Microbiology and Biotechnology (2022)
-
The critical role that spectral libraries play in capturing the metabolomics community knowledge
Metabolomics (2022)
-
Cognitive analysis of metabolomics data for systems biology
Nature Protocols (2021)