Abstract
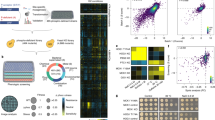
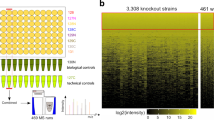
Yeast libraries revolutionized the systematic study of cell biology. To extensively increase the number of such libraries, we used our previously devised SWAp-Tag (SWAT) approach to construct a genome-wide library of ~5,500 strains carrying the SWAT NOP1promoter-GFP module at the N terminus of proteins. In addition, we created six diverse libraries that restored the native regulation, created an overexpression library with a Cherry tag, or enabled protein complementation assays from two fragments of an enzyme or fluorophore. We developed methods utilizing these SWAT collections to systematically characterize the yeast proteome for protein abundance, localization, topology, and interactions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
02 January 2019
The version of Supplementary Table 1 originally published online with this article contained incorrect localization annotations for one plate. This error has been corrected in the online Supplementary Information.
References
Botstein, D. & Fink, G. R. Yeast: an experimental organism for 21st century biology. Genetics 189, 695–704 (2011).
Giaever, G. et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 (2002).
Huh, W.-K. et al. Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003).
Tarassov, K. et al. An in vivo map of the yeast protein interactome. Science 320, 1465–1470 (2008).
Yofe, I. et al. One library to make them all: streamlining the creation of yeast libraries via a SWAp-Tag strategy. Nat. Methods 13, 371–378 (2016).
Khmelinskii, A., Meurer, M., Duishoev, N., Delhomme, N. & Knop, M. Seamless gene tagging by endonuclease-driven homologous recombination. PLoS One 6, e23794 (2011).
Meurer, M. et al. A genome-wide resource for high-throughput genomic tagging of yeast ORFs. bioRxiv Preprint at https://www.biorxiv.org/content/early/2017/11/30/226811 (2017).
Engel, S. R. & Cherry, J. M. The new modern era of yeast genomics: community sequencing and the resulting annotation of multiple Saccharomyces cerevisiae strains at the Saccharomyces Genome Database. Database (Oxf.) 2013, bat012 (2013).
Pédelacq, J.-D., Cabantous, S., Tran, T., Terwilliger, T. C. & Waldo, G. S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 (2006).
Mumberg, D., Müller, R. & Funk, M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122 (1995).
Sun, J. et al. Cloning and characterization of a panel of constitutive promoters for applications in pathway engineering in Saccharomyces cerevisiae. Biotechnol. Bioeng. 109, 2082–2092 (2012).
Breker, M., Gymrek, M. & Schuldiner, M. A novel single-cell screening platform reveals proteome plasticity during yeast stress responses. J. Cell Biol. 200, 839–850 (2013).
Picotti, P. et al. A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature 494, 266–270 (2013).
Weinberg, D. E. et al. Improved ribosome-footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Rep. 14, 1787–1799 (2016).
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. S. & Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009).
Neymotin, B., Athanasiadou, R. & Gresham, D. Determination of in vivo RNA kinetics using RATE-seq. RNA 20, 1645–1652 (2014).
Belle, A., Tanay, A., Bitincka, L., Shamir, R. & O’Shea, E. K. Quantification of protein half-lives in the budding yeast proteome. Proc. Natl. Acad. Sci. USA 103, 13004–13009 (2006).
Chen, X. & Zhang, J. The genomic landscape of position effects on protein expression level and noise in yeast. Cell Syst. 2, 347–354 (2016).
Weill, U. et al. Toolbox: creating a systematic database of secretory pathway proteins uncovers new cargo for COPI. Traffic 19, 370–379 (2018).
Morgenstern, M. et al. Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Rep. 19, 2836–2852 (2017).
Fukasawa, Y. et al. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell. Proteom. 14, 1113–1126 (2015).
Emanuelsson, O., Brunak, S., von Heijne, G. & Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971 (2007).
Vögtle, F.-N. et al. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 139, 428–439 (2009).
Venne, A. S., Vögtle, F.-N., Meisinger, C., Sickmann, A. & Zahedi, R. P. Novel highly sensitive, specific, and straightforward strategy for comprehensive N-terminal proteomics reveals unknown substrates of the mitochondrial peptidase Icp55. J. Proteome Res. 12, 3823–3830 (2013).
Backes, S. et al. Tom70 enhances mitochondrial preprotein import efficiency by binding to internal targeting sequences. J. Cell Biol. 217, 1369–1382 (2018).
Chacinska, A. et al. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 23, 3735–3746 (2004).
Wiedemann, N. et al. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature 424, 565–571 (2003).
Ben-Menachem, R. & Pines, O. Detection of dual targeting and dual function of mitochondrial proteins in yeast. in Mitochondria: Practical Protocols (eds. Mokranjac, D. & Perocchi, F.) 179–195 (Springer, New York, 2017).
Eisenberg-Bord, M. & Schuldiner, M. Mitochatting: if only we could be a fly on the cell wall. Biochim. Biophys. Acta 1864, 1469–1480 (2017).
Eisenberg-Bord, M. & Schuldiner, M. Ground control to major TOM: mitochondria-nucleus communication. FEBS J. 284, 196–210 (2017).
Jin, L. et al. Random insertion of split-cans of the fluorescent protein Venus into Shaker channels yields voltage sensitive probes with improved membrane localization in mammalian cells. J. Neurosci. Methods 199, 1–9 (2011).
Erdmann, R. Assembly, maintenance and dynamics of peroxisomes. Biochim. Biophys. Acta 1863, 787–789 (2016).
Kim, H., Melén, K., Osterberg, M. & von Heijne, G. A global topology map of the Saccharomyces cerevisiae membrane proteome. Proc. Natl. Acad. Sci. USA 103, 11142–11147 (2006).
Kivioja, T. et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods 9, 72–74 (2011).
Douglas, A. C. et al. Functional analysis with a barcoder yeast gene overexpression system. G3 (Bethesda) 2, 1279–1289 (2012).
Janke, C. et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 (2004).
Yofe, I. & Schuldiner, M. Primers-4-Yeast: a comprehensive web tool for planning primers for Saccharomyces cerevisiae. Yeast 31, 77–80 (2014).
Brachmann, C. B. et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 (1998).
Gietz, R. D. & Woods, R. A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350, 87–96 (2002).
Sopko, R. et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell 21, 319–330 (2006).
Hin Yan Tong, A. & Boone, C. High-throughput strain construction and systematic synthetic lethal screening in Saccharomyces cerevisiae. Methods Mol. Biol. 36, 1–19 (2007).
Cohen, Y. & Schuldiner, M. Advanced methods for high-throughput microscopy screening of genetically modified yeast libraries. Methods Mol. Biol. 781, 127–159 (2011).
Knox, C., Sass, E., Neupert, W. & Pines, O. Import into mitochondria, folding and retrograde movement of fumarase in yeast. J. Biol. Chem. 273, 25587–25593 (1998).
Weckbecker, D., Longen, S., Riemer, J. & Herrmann, J. M. Atp23 biogenesis reveals a chaperone-like folding activity of Mia40 in the IMS of mitochondria. EMBO J. 31, 4348–4358 (2012).
Goldstein, A. L. & McCusker, J. H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541–1553 (1999).
Flagfeldt, D. B., Siewers, V., Huang, L. & Nielsen, J. Characterization of chromosomal integration sites for heterologous gene expression in Saccharomyces cerevisiae. Yeast 26, 545–551 (2009).
Stark, C. et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 34, D535–D539 (2006).
Harper, J. W., Adami, G. R., Wei, N., Keyomarsi, K. & Elledge, S. J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75, 805–816 (1993).
Bartel, P., Chien, C. T., Sternglanz, R. & Fields, S. Elimination of false positives that arise in using the two-hybrid system. Biotechniques 14, 920–924 (1993).
Knoblach, B. et al. An ER-peroxisome tether exerts peroxisome population control in yeast. EMBO J. 32, 2439–2453 (2013).
Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 (2001).
Tusnády, G. E. & Simon, I. The HMMTOP transmembrane topology prediction server. Bioinformatics 17, 849–850 (2001).
Käll, L., Krogh, A. & Sonnhammer, E. L. L. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res. 35, 429–432 (2007).
Reynolds, S. M., Käll, L., Riffle, M. E., Bilmes, J. A. & Noble, W. S. Transmembrane topology and signal peptide prediction using dynamic Bayesian networks. PLOS Comput. Biol. 4, e1000213 (2008).
Bernsel, A., Viklund, H., Hennerdal, A. & Elofsson, A. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 37, 465–468 (2009).
Acknowledgements
We thank Y. Peleg for plasmid construction, G. Brodsky for graphics, R. Rotkopf for support in statistical analysis, K. Tedrick for technical help with the Y2H experiments, and C. Meisinger and N. Vögtle for help with the MTS assignments. We thank G. Krieger for helpful discussions and technical help. We thank C. Ungermann (University of Osnabrück, Germany) and W.-K. Huh (Seoul National University, Seoul, South Korea) for plasmids. The work in the Schuldiner laboratory was supported by ERC CoG Peroxisystem (646604), SFB 1190 from the DFG, a Mitzutani foundation grant, and a VolksWagen foundation grant (93092). The collaborative work on this manuscript done by the Schuldiner, Pines, Herrmann, and Rapaport laboratories was supported by a DIP grant (P17516). Work at the Rachubinski lab was supported by Foundation Grant FDN-143289 from the Canadian Institutes of Health Research. Work in the Michnick lab was supported by Canadian Institutes of Health Research grant MOP-GMX-152556. Work in the Levy lab was supported by Israel Science Foundation grants 1775/12 and 2179/14. U.W. and D.D. are recipients of the Azrieli student-award grant. M.S. is an Incumbent of the Dr. Gilbert Omenn and Martha Darling Professorial Chair in Molecular Genetics.
Author information
Authors and Affiliations
Contributions
U.W., I.Y., and M.S. conceived the study. U.W., I.Y., E.S., B.S., D.D., J.N., R.B.M., Z.A., O.G., N.H., S.C., K.K., B.K., J.L., F.B., J.K., and S.B.-D. carried out the investigation. M.S. and U.W. wrote the manuscript. All of the authors reviewed and edited the manuscript. E.Z., J.M.H., R.A.R., O.P., D.R., S.W.M., E.D.L., and M.S. supervised the work and acquired funding.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Conditionally expressed proteins regain metabolic regulation when tagged in a native promoter/regulation manner.
(a) NOP1pr-GFP-GAL2 yeast strain before and after native promoter/regulation GFP swap to a NATIVEpr-GFP tag in synthetic medium with either 2% glucose or 2% galactose. (b) NOP1pr-GFP-PHO5 yeast strain before and after native promoter/regulation GFP swap to a NATIVEpr-GFP tag in synthetic medium with or depleted of phosphate. (c) NOP1pr-GFP-SUC2 yeast strain before and after native promoter/regulation GFP swap to a NATIVEpr-GFP tag in synthetic medium with either 2% or 0.2% glucose. All scale bars are 5µm. Imaging of strains was performed a single time. Images represent entire field.
Supplementary Figure 2 Genome-wide N′-tagged collections’ signal intensity distribution.
(a) Histogram of the expression levels of fluorophore-protein fusions of the TEF2pr-mCherry, NATIVEpr-GFP and NOP1pr-GFP libraries. a.u., arbitrary units. (b) Scatter plots showing the correlation between protein abundance of NATIVEpr-GFP tagged strains versus C’ GFP tagged strains12. a.u., arbitrary units. R represents two-sided Spearman correlation test score. Quantitation of abundance was preformed once. Strains with a final abundance score lower than 1, were excluded from the data analysis and Spearman correlation tests.
Supplementary Figure 3 N′ SWAT tagging of proteins bearing an MTS.
(a) Two MTS-containing proteins, Tuf1 and Coq10, were N’-tagged with and without a generic MTS in the tagging cassette. (b) Native promoter/regulation swapping of N’ SWAT-tagged Tuf1 restores the native MTS targeting to mitochondria. All scale bars are 5µm. Imaging of strains was performed a single time. Images represent entire field. (c) Mitochondrial proteins tagged with a TEF2pr-mCherry without an MTS (d) MTS and TMD prediction analysis for proteins showing mitochondrial localization with the NOP1pr-GFP, NATIVEpr-GFP, and/or C’ GFP tags. Cx(9)C - cysteine-rich domain26, β-barrel domain27. All scale bars are 5µm. Imaging of strains was performed a single time. Images represent entire field.
Supplementary Figure 4 Ysa1 is dual-localized to mitochondria.
(a) N’-tagged Ysa1 under its native promoter has a fraction that co-localizes with the mitochondrial marker Tom20-mCherry. (b) C’-tagged Ysa1 has a fraction that co-localizes with the mitochondrial marker Tom20-mCherry. (c) α-tagged Ysa1 is dually localized between mitochondria (M) and cytosol (C) in subcellular fractionation. Total lysate (T) is shown as control. Hsp60 is probed as a mitochondrial protein. Hxk1 is probed as a cytosolic protein. All scale bars are 5µm. Imaging of strains was performed a single time. Images represent entire field.
Supplementary Figure 5 Tam41 can be imported into isolated mitochondria in the absence of its MTS.
The proteins Tam41 (a) and Coq2 (b) and their N-terminally truncated versions (∆MTS) were synthesized in the presence of 35S-methionine in reticulocyte lysate. The 35S-methionine-radiolabeled proteins were then incubated with wild type mitochondria for the indicated times. Non-imported protein was removed by proteinase K (PK) treatment. 10% of the protein used per lane was loaded as control. In the samples labeled with VAO, the membrane potential of mitochondria was dissipated before radiolabeled proteins were added. Positions of precursor (pre) and mature (mat) forms are indicated. The graphs show targetP scores of internal matrix targeting sequence-like regions for each residue in the sequences of Coq2 and Tam4125. The blue dashed line marks the processing site in the proteins for MPP, which removes their N-terminal targeting sequences. The nine green boxes indicate the positions of the transmembrane domains of Coq2.
Supplementary Figure 6 Mitochondrial proteins that are differentially localized depending on medium conditions.
NATIVEpr-GFP tagged mitochondrial proteins suspected to be dually localized were visualized under different medium conditions. Cells were grown in glucose 2% mid-log, glycerol 2% mid-log, galactose 2% mid-log, glucose 0.2% mid-log, or glucose 2% stationary for 4 hours before imaging. All scale bars are 5µm. Imaging of strains was performed a single time. Images represent entire field.
Supplementary Figure 7 Pex17 and Inp1 interact in a yeast two-hybrid assay.
S. cerevisiae HF7c cells expressing Gal4-AD and Gal4-BD protein fusions to Pex17 and Inp1 were tested for the ability of the protein fusions to interact by a yeast two-hybrid growth assay. A minus sign (-) denotes empty vector. A serial dilution series was spotted onto -Leu -Trp medium (left) and -His -Leu -Trp medium (right). Growth on -Leu -Trp medium requires the presence of both AD and BD plasmids in cells and is indicative of cell density. Growth on -His -Leu -Trp medium occurs only in the presence of a protein-protein interaction.
Supplementary Figure 8 Scm4 is an outer-membrane mitochondrial protein with both its N′ and C′ termini facing the cytosol.
(a) TEF2pr-VC-SCM4 with a cytosolic-VN co-localizes with the mitochondrial marker MTS-BFP. (b) Proteinase K (PK) protection assay verifies that Scm4 is an outer membrane mitochondrial protein with both its N’ and C’ termini facing the cytosol. Mitochondria were isolated from cells expressing Scm4 (~21 kDa) HA-tagged at either the N’ (HA-Scm4) or C’ (Scm4-HA). Isolated organelles were treated with the indicated amounts of either PK or trypsin. In some samples the detergent Triton X‐100 was added (+TX-100) or the outer membrane of the organelles was ruptured by swelling (+swelling). Immunodecoration was carried out with antibodies against the HA‐tag, the outer membrane protein Tom70 (70 kDa) that is exposed to the cytosol, or the matrix protein Aco1 (90 kDa). Scale bar is 5µm. Imaging of strains was performed a single time. Images represent entire field.
Supplementary Figure 9 The TEF2pr-VC tag with a cytosolic VN helps to determine the topology for membrane-spanning proteins.
Membrane-spanning proteins that showed a signal with the TEF2pr-VC tag and cytosolic VN can be considered as having their N’ facing the cytosol (“in”). C’ topology from33. Program predictions for number of TMDs and N’ topology. NA – Not Available. Scale bar is 5µm. Imaging of strains was performed a single time. Images represent entire field.
Supplementary Figure 10 Full scans of all blots.
(a-c) Original blots for supplementary figure 3c. b and c are the same blots in different exposure times. Black boxes mark the areas used for the figure. The relevant antibody for the boxes is underlined. (d and e) Original blots for supplementary figures 4a and 4b. (f and g) Original blots for supplementary figure 7b.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1-10
Supplementary Table 1
All information on strains in the genome-wide SWAT library
Supplementary Table 2
Mitochondrial targeting sequence (MTS) predictions
Supplementary Table 3
Check PCR and Anchor Seq analysis
Supplementary Table 4
Results of peroxisomal protein interaction screen using DHFR PCA
Supplementary Table 5
Results of peroxisomal protein interaction screen using Split Venus PCA
Supplementary Table 6
Topology assignments
Supplementary Table 7
All plasmids used in this study
Supplementary Table 8
All primers used in this study
Supplementary Table 9
Genes that could not be N-terminally tagged with the SWAT cassettes
Rights and permissions
About this article
Cite this article
Weill, U., Yofe, I., Sass, E. et al. Genome-wide SWAp-Tag yeast libraries for proteome exploration. Nat Methods 15, 617–622 (2018). https://doi.org/10.1038/s41592-018-0044-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-018-0044-9
This article is cited by
-
From beer to breadboards: yeast as a force for biological innovation
Genome Biology (2024)
-
MemPrep, a new technology for isolating organellar membranes provides fingerprints of lipid bilayer stress
The EMBO Journal (2024)
-
The social and structural architecture of the yeast protein interactome
Nature (2023)
-
Orphan quality control by an SCF ubiquitin ligase directed to pervasive C-degrons
Nature Communications (2023)
-
The vacuolar iron transporter mediates iron detoxification in Toxoplasma gondii
Nature Communications (2023)