Abstract
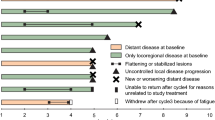
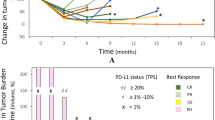
Talimogene laherparepvec (T-VEC) is an oncolytic virus hypothesized to enhance triple-negative breast cancer (TNBC) responses to neoadjuvant chemotherapy (NAC). This article describes the phase 2 trial of T-VEC plus NAC (ClinicalTrials.gov ID: NCT02779855). Patients with stage 2–3 TNBC received five intratumoral T-VEC injections with paclitaxel followed by doxorubicin and cyclophosphamide and surgery to assess residual cancer burden index (RCB). The primary end point was RCB0 rate. Secondary end points were RCB0–1 rate, recurrence rate, toxicity and immune correlates. Thirty-seven patients were evaluated. Common T-VEC toxicities were fevers, chills, headache, fatigue and injection site pain. NAC toxicities were as expected. Four thromboembolic events occurred. The primary end point was met with an estimated RCB0 rate = 45.9% and RCB0–1 descriptive rate = 65%. The 2-year disease-free rate is equal to 89% with no recurrences in RCB0–1 patients. Immune activation during treatment correlated with response. T-VEC plus NAC in TNBC may increase RCB0–1 rates. These results support continued investigation of T-VEC plus NAC for TNBC.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The RNAseq dataset is available on dbGAP (https://dbgap.ncbi.nlm.nih.gov/) to researchers with an approved dbGAP profile as outlined on the website, using the accession number phs003199.v1.p1. All other datasets generated during and/or analysed during the current study can be requested from the corresponding author. Requests will be responded to within 4 weeks.
Change history
17 March 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41591-023-02309-4
References
Bianchini, G., De Angelis, C., Licata, L. & Gianni, L. Treatment landscape of triple-negative breast cancer—expanded options, evolving needs. Nat. Rev. Clin. Oncol. 19, 91–113 (2022).
Zhu, W. et al. Age-related disparity in immediate prognosis of patients with triple-negative breast cancer: a population-based study from SEER cancer registries. PLoS ONE 10, e0128345 (2015).
Miyashita, M. et al. Tumor-infiltrating CD8+ and FOXP3+ lymphocytes in triple-negative breast cancer: its correlation with pathological complete response to neoadjuvant chemotherapy. Breast Cancer Res. Treat. 148, 525–534 (2014).
Heise, C. et al. ONYX-015, an E1B gene-attenuated adenovirus, causes tumor-specific cytolysis and antitumoral efficacy that can be augmented by standard chemotherapeutic agents. Nat. Med. 3, 639–645 (1997).
Soliman, H. et al. A phase I trial of talimogene laherparepvec in combination with neoadjuvant chemotherapy for the treatment of nonmetastatic triple-negative breast cancer. Clin. Cancer Res. 27, 1012–1018 (2021).
Schmid, P. et al. Pembrolizumab for early triple-negative breast cancer. N. Engl. J. Med. 382, 810–821 (2020).
Schmid, P. et al. Event-free survival with pembrolizumab in early triple-negative breast cancer. N. Engl. J. Med. 386, 556–567 (2022).
Ribas, A. et al. MASTERKEY-265: a phase III, randomized, placebo (Pbo)-controlled study of talimogene laherparepvec (T) plus pembrolizumab (P) for unresectable stage IIIB-IVM1c melanoma (MEL). Ann. Oncol. 32, S868–S869 (2021).
Symmans, W. F. et al. Assessment of residual cancer burden and event-free survival in neoadjuvant treatment for high-risk breast cancer: an analysis of data from the I-SPY2 randomized clinical trial. JAMA Oncol. 7, 1654–1663 (2021).
Yau, C. et al. Residual cancer burden after neoadjuvant chemotherapy and long-term survival outcomes in breast cancer: a multicentre pooled analysis of 5161 patients. Lancet Oncol. 23, 149–160 (2022).
Park, Y. H. et al. Chemotherapy induces dynamic immune responses in breast cancers that impact treatment outcome. Nat. Commun. 11, 6175 (2020).
Loibl, S. et al. A randomised phase II study investigating durvalumab in addition to an anthracycline taxane-based neoadjuvant therapy in early triple-negative breast cancer: clinical results and biomarker analysis of GeparNuevo study. Ann. Oncol. 30, 1279–1288 (2019).
Gaya, M. et al. Initiation of antiviral B cell immunity relies on innate signals from spatially positioned NKT cells. Cell 172, 517–533 (2018).
Gebremeskel, S. et al. Natural killer T cell immunotherapy combined with oncolytic vesicular stomatitis virus or reovirus treatments differentially increases survival in mouse models of ovarian and breast cancer metastasis. J. Immunother. Cancer 9, e002096 (2021).
Kaufman, H. L., Kohlhapp, F. J. & Zloza, A. Oncolytic viruses: a new class of immunotherapy drugs. Nat. Rev. Drug Discov. 15, 660 (2016).
Kaur, B., Mukhlis, Y., Natesh, J., Penta, D. & Musthapa Meeran, S. Identification of hub genes associated with EMT-induced chemoresistance in breast cancer using integrated bioinformatics analysis. Gene 809, 146016 (2022).
Zhu, Z. et al. Expression of AQP3 and AQP5 as a prognostic marker in triple-negative breast cancer. Oncol. Lett. 16, 2661–2667 (2018).
Salgado, R. et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann. Oncol. 26, 259–271 (2015).
Bushnell, B., Rood, J. & Singer, E. BBMerge—accurate paired shotgun read merging via overlap. PLoS ONE 12, e0185056 (2017).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Aran, D., Hu, Z. & Butte, A. J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 18, 220 (2017).
Chen, X. et al. TNBCtype: a subtyping tool for triple-negative breast cancer. Cancer Inform. 11, 147–156 (2012).
Acknowledgements
This work was supported by the digital pathology, tissue processing, molecular genomics, bioinformatics/biostatistics and flow cytometry cores at the Moffitt Cancer Center funded by a Moffitt Cancer Center support grant no. P30-CA076292. We thank A. Aldrich for the flow cytometry data acquisition.
Author information
Authors and Affiliations
Contributions
H.S. was responsible for study design and oversight, protocol and manuscript authorship, data acquisition, data interpretation and manuscript approval. H.K., M.C.L., H.H., M.R., A.W., D.H., S.F., A.S., N.K., J.K., C.L., A.A., R.C., R.J.W., B.M., A.C., B.N., S.H. and B.C. were responsible for study design, data acquisition and data interpretation, and manuscript critical review, editing and approval.
Corresponding author
Ethics declarations
Competing interests
Funding for the trial was provided by Amgen. H.S. has received consulting fees from AstraZeneca, Novartis, Seattle Genetics, PUMA, Sanofi and Eisai; licensing fees for intellectual property from Celyad Oncology; and institutional research funding from Amgen. H.K. has sat on the scientific advisory board for Celcuity; holds equities in Agenus, TG Therapeutics, Vexart, Lipocine, Mustang Bio, MEI Pharma and Tiziana Life Sciences; and has received institutional research support from AstraZeneca. M.C.L. has received research funding from Elucent Medical. S.H. has sat on the advisory board of Devicor/Mammotome. H.H. has received fees for advisory and speaker bureau activities from Lilly, Novartis and Immunomedics. B.C. has received advisory fees from Merit Oncology and has intellectual property rights with Immunorestoration. M.R. has received speaker fees from Roche. A.W., D.H., S.F., A.S., N.K., J.K., C.L., A.A., R.C., R.J.W., B.M. and A.C. declare no competing interests.
Peer review
Peer review information
Nature Medicine thanks S.-H. Jung, H. Kaufman, M. Kok and A. van Akkooi for their contribution to the peer review of this work. Primary Handling Editor: Ulrike Harjes, in collaboration with the Nature Medicine team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 OptSNE analysis of baseline circulating lymphocytes.
The image shows the identified lymphocyte clusters of closely related populations along with their dominant cell surface markers. The first panel shows which clusters of cells are from responders (events in red) versus non-responders (events in yellow). Four clusters highlighted in the figure are particularly enriched for events from responders. The first three clusters appear to be natural killer T cell subsets (NKT) and the fourth cluster a B cell predominant one.
Extended Data Fig. 2 Gene expression associated with poor response.
Volcano plot showing statistically significant differentially expressed individual genes represented by red dots.
Extended Data Fig. 3 Gating illustration.
A representative ancestry gating figure for the various populations quantified.
Supplementary information
Supplementary Information
Study protocol document.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Soliman, H., Hogue, D., Han, H. et al. Oncolytic T-VEC virotherapy plus neoadjuvant chemotherapy in nonmetastatic triple-negative breast cancer: a phase 2 trial. Nat Med 29, 450–457 (2023). https://doi.org/10.1038/s41591-023-02210-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-023-02210-0
This article is cited by
-
Therapeutic cancer vaccines: advancements, challenges, and prospects
Signal Transduction and Targeted Therapy (2023)
-
Exploiting Therapeutic Vulnerabilities in Triple-Negative Breast Cancer: Successes, Challenges, and Opportunities
Current Breast Cancer Reports (2023)