Abstract
Conserved epitopes on the influenza hemagglutinin (HA) stem are an attractive target for universal vaccine strategies as they elicit broadly neutralizing antibodies. Such antibody responses to stem-specific epitopes have been extensively characterized for HA subtypes H1 and H5 in humans. H2N2 influenza virus circulated 50 years ago and represents a pandemic threat due to the lack of widespread immunity, but, unlike H1 and H5, the H2 HA stem contains Phe45HA2 predicted to sterically clash with HA stem-binding antibodies characterized to date. To understand the effect of Phe45HA2, we compared the HA stem-specific B cell response in post hoc analyses of two phase 1 clinical trials, one testing vaccination with an H2 ferritin nanoparticle immunogen (NCT03186781) and one with an inactivated H5N1 vaccine (NCT01086657). In H2-naive individuals, the magnitude of the B cell response was equivalent, but H2-elicited HA stem-binding B cells displayed greater cross-reactivity than those elicited by H5. However, in individuals with childhood H2 exposure, H5-elicited HA stem-binding B cells also displayed high cross-reactivity, suggesting recall of memory B cells formed 50 years ago. Overall, we propose that a one-residue difference on an HA immunogen can alter establishment and expansion of broadly neutralizing memory B cells. These data have implications for stem-based universal influenza vaccination strategies.
Similar content being viewed by others
Main
Studies estimating the worldwide influenza-associated mortality between 1999 and 2015 show that yearly influenza virus epidemics are associated with 290,000–650,000 deaths annually1. H1N1, H3N2 and H2N2 influenza subtypes have caused pandemics over the past 100 years, including the 1918 H1N1 influenza pandemic, which led to 20–50 million deaths. H2N2 influenza virus entered the human population in 1957 through reassortment with avian H2N2 and human H1N1, causing a pandemic that led to 1–4 million deaths worldwide. H2N2 stopped circulating in the human population in 1968 when a reassortment event with H3 viruses replaced it with human H3N2.
HA, comprising a head and stem domain, is the most abundant glycoprotein on the influenza virus surface and can be divided phylogenetically into 18 subtypes, which segregate into two major groups: group 1 and group 2. Protection against influenza infection correlates with titers of HA-binding antibodies that block viral attachment and infection of cells. Current yearly seasonal influenza vaccines primarily induce an antibody response directed against the immunodominant but continually diversifying HA head region. These antibody responses provide protection against the vaccinating strain but little cross-protection against other strains or subtypes. As a result, there has been a concerted effort to develop immunogens that elicit broadly neutralizing immune responses focused on subdominant, but more conserved, epitopes, such as those on the HA stem domain, that might protect against viruses of pandemic threat, including H2, H5, H6 and H9 subtypes.
In response to H1 or H5 viral infection or vaccination, most HA stem-specific responses comprise a convergent class of immunoglobulins with VH1-69 heavy chains capable of broadly neutralizing group 1 HA subtypes2,3,4,5,6,7,8,9,10, but the B cell response to the H2 HA stem domain has not been evaluated. Human H2N2 influenza strains are the only strains to have a bulky phenylalanine in HA at residue 45HA2 located near the hydrophobic pocket targeted by VH1-69-class antibodies, and this residue is predicted to sterically clash with these antibodies11. Indeed, several broadly binding HA stem-binding monoclonal antibodies (mAbs) have selectively low binding to H2 HA6,12,13,14,15,16, and introduction of a glycan at residue 45HA2 knocks out most HA stem binding17,18. In light of the central role of HA residue 45HA2 in antibody binding, we hypothesized that the nature of the HA stem-binding immunoglobulin repertoire induced by H2 HA might be different than that induced by other group 1 HAs.
To evaluate the effect of Phe45HA2 on H2 HA-specific humoral immune responses, we compared the magnitude and cross-reactivity of the HA stem-binding cellular and serological B cell responses elicited by H5 versus H2 vaccination. Altogether, we conclude that presence of the bulky phenylalanine at residue 45HA2 on H2 HA selects for B cells with greater breadth of immunoglobulin binding and neutralization across group 1 subtypes. Moreover, analysis of the HA stem response in individuals exposed to H2 50 years ago suggests that an H2 immunogen can lead to a long-term HA stem-specific memory repertoire capable of providing broad protection across the lifetime of an individual. Based on these findings, we propose that incorporating H2 instead of H1 or H5 in an HA stem-based vaccine design might provide broader protection against potential group 1 pandemic threats.
Results
Study design
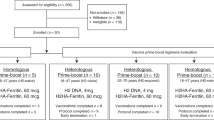
Unlike most other influenza subtypes, the HA proteins from human H2N2 influenza strains have a phenylalanine at 45HA2 predicted to sterically clash with many HA stem-binding mAbs described to date, including VH1-69-class mAbs, which dominate the group 1 HA stem-specific response (Fig. 1a,b)16. To test the effect of Phe45HA2 on H2 HA in humans, we compared the HA stem-specific memory B cell response upon vaccination with H5 versus H2 in individuals naive to both HA subtypes in two phase 1 clinical trials. In the first trial (NCT01086657), individuals were primed with A/Indonesia/05/2005 H5 DNA or matched H5N1 monovalent inactivated vaccine (H5 protein), followed by a boost with the H5N1 monovalent inactivated vaccine administered 16–24 weeks later (Fig. 1c)19. In the second trial (NCT03186781), individuals were primed with A/Singapore/2/1957 H2 DNA or an H2 HA ferritin nanoparticle (H2 protein) vaccine, followed by a boost with the H2 HA ferritin nanoparticle vaccine 16 weeks later (Fig. 1c).
a, Comparison of residues in HA2 proteins from different group 1 and group 2 subtypes as indicated. Residue 45HA2 and H2N2 strains are highlighted with red boxes. HA strains shown are the following: A/Brevig Mission/1/1918 (H1N1 BM18), A/NewCaledonia/20/1999 (H1N1 NC99), A/California/04/2009 (H1N1 CA09), A/Singapore/1/1957 (H2N2 SI57), A/AnnArbor/6/1960 (H2N2 AA60), A/Canada/720/2005 (H2N2 CAN05), A/Indonesia/05/2005 (H5N1 IN05), A/Taiwan/1/2013 (H6N1 TW13), A/Guangxi-Xiangshan/11522/2018 (H9N2 GX18), A/Perth/16/2009 (H3N2 PE09), A/Shanghai/2/2013 (H7N9 SH13) and A/Jiangxi-Donghu/346/2013 (H10N8 JD13). Strains in bold were used for the vaccine trials described in c. b, Ribbon diagrams showing the crystal structure of mAb CR6261 in complex with H1 A/Brevig Mission/1/1918 HA (PDB, 3GBN) with the wild-type Ile45 residue (top) or modeled with Phe45 instead (bottom). CR6261 is colored purple; HA is colored teal or gray. Residue clashes are illustrated with red plates. c, Schematic showing vaccine regimens and time points analyzed for the H5 and H2 phase 1 vaccine trials. H5 protein is from a monovalent influenza subunit virion (MIV), and H2 protein is from an H2 HA ferritin nanoparticle. W, week.
H2 vaccination elicits memory B cells with greater breadth
We first used flow cytometry to determine the magnitude and cross-reactivity of the HA stem-specific memory B cell response in 15 individuals in each trial 2 weeks after the vaccine boost (Fig. 1c). H5 has never circulated in humans in North America, and we limited analysis to individuals from each trial who were born after 1970 to ensure that all individuals were naive to both H5 and H2 (Supplementary Table 1). We gated on CD19+CD20+IgG+ memory B cells and used a combination of fluorescently labeled HA trimers and stabilized HA stem-only trimers to detect memory B cells able to bind the stem region of H1, H2 or H5 HA (Fig. 2a and Supplementary Fig. 1a)10,20,21. Although responses were variable, little evidence of a difference in the total vaccine-specific memory B cell HA response was observed between trials, whether individuals were primed with DNA or protein (Fig. 2b). The proportion of the memory B cell HA vaccine response that recognized the HA stem region was also not significantly different between the two vaccine trials, although it varied with the prime (Fig. 2c). DNA-primed individuals in both trials had a higher proportion of HA stem-binding B cells than protein-primed individuals (Fig. 2c). We next compared the cross-reactivity of the HA stem-specific responses by determining the percentage of HA stem-specific memory B cells that cross-reacted with the H1 HA stem domain and the vaccinating HA (H2 or H5) or bound all three (H1, H2 and H5) (Fig. 2a). We limited the analysis to group 1 subtypes as we observed very little cross-reactivity to group 2 HA subtypes (Supplementary Fig. 1b). Within the population negative for binding to the H1 HA stem domain, we also looked for cells that bound the H2 and/or H5 stem domain and found that almost all HA stem-specific B cells elicited by H2 or H5 vaccination were cross-reactive with H1 (Fig. 2d). Interestingly, whether individuals were primed by DNA or protein, the proportion of stem-specific memory B cells that bound all three subtypes—H1, H2 and H5—was significantly higher after H2 vaccination than after H5 vaccination (Fig. 2d). Although, on average, over 85% of HA stem-specific memory B cells generated by H2 vaccination also cross-reacted with H5, only about 65% of HA stem-specific memory B cells elicited by H5 vaccination also bound H2 in addition to H1 (Fig. 2d). This difference in cross-reactivity was unique to HA stem-specific memory B cells. We detected no discernable difference in the cross-reactivity of HA head-binding memory B cells between the two trials (Supplementary Fig. 1c,d). To test whether Phe45HA2 on the H2 HA stem was responsible for reduced cross-reactivity to H2 after H5 vaccination, we mutated Phe45 on H2 to isoleucine (H2 F45I) to match H5. Indeed, close to 90% of H5 vaccine-elicited HA stem-binding memory B cells could bind the H2 F45I-stabilized stem mutant (Fig. 2e). Overall, these data indicate that vaccination with H5 HA elicits memory B cells with varied ability to bind H2 HA, whereas H2 vaccination preferentially expands memory B cells capable of recognizing H2, H5 and H1 HAs.
a, PBMCs isolated 2 weeks after boost after either H5 (top) or H2 (bottom) vaccination were stained with antibodies to detect B cell surface markers and fluorescently labeled HA probes. CD19+CD20+IgG+ B cells were gated, and the reactivity of memory B cells to H2 HA, H5 HA, H1 stem, H5 stem and H2 stem was determined as indicated. b, The percentage of CD19+CD20+IgG+ B cells that bound the vaccinating HA—H5 HA (H5+) after H5 vaccination and H2 HA (H2+) after H2 vaccination—2 weeks after the vaccine boost. Individuals are divided according to whether they were primed with a DNA vaccine (left) or a protein vaccine (right). c, The proportion of B cells binding H5 HA or H2 HA that also bound the corresponding HA stem. d, The proportion of B cells binding the HA stem that were triple reactive, binding H1H5H2; were double reactive, binding H1H5; H1H2 or H5H2; or were single reactive, binding H5 or H2 only. Data are shown separately for individuals who were primed with a DNA vaccine (left) or a protein vaccine (right). e, Proportion of HA stem-binding B cells from each vaccine study at the same time point that were cross-reactive with H5, H1 and either wild-type (WT) H2 or the F45I H2 stem mutant as indicated. For b–e, each dot represents one individual, with the line indicating the mean. n = 15 individuals vaccinated with H2 (eight DNA prime and seven protein prime) and 15 individuals vaccinated with H5 (11 DNA prime and four protein prime). Data are representative of two independent experiments using different combinations of HA probes. Statistical significance was determined for b–d using the two-sided Mann–Whitney test and for e using the two-sided Wilcoxon matched-pairs rank test.
H2 vaccination results in greater serological breadth
To see whether this difference in memory B cell reactivity corresponds to differences in the antibody serological response, we compared the H2 and H5 serological binding and neutralization levels in H2- and H5-vaccinated individuals 2 weeks after the vaccine boost. Total H2 and H5 serological binding to the vaccinating subtype was not significantly different between the two trials (P = 0.9, Mann–Whitney test) (Fig. 3a). Serological binding to the vaccinating HA was highest in each vaccine group, but the difference between H2 and H5 binding in H2-vaccinated individuals was smaller than for H5-vaccinated individuals (Fig. 3a,b). When we measured H2 and H5 neutralization titers, we saw a similar trend. Although H5 neutralization titers were only slightly lower than H2 neutralization titers in H2-vaccinated individuals, H2 neutralization titers were substantially lower in H5-vaccinated individuals (Fig. 3c,d). We also noted that H2 neutralization titers were lower upon H2 vaccination than H5 neutralization titers in H5-vaccinated individuals (Fig. 3c), possibly due to differences in neutralization sensitivity between the two viruses, as we observed high H2 binding titers in H2 vaccinees (Fig. 3a). Both HA head- and stem-specific antibodies can contribute to serological neutralization, so, to evaluate HA stem-specific responses, we measured serological H2, H5 and H1 HA stem binding with the same stabilized stem probes used in flow cytometry. Although we detected a difference between H2 and H5 stem binding in both trials (Fig. 3e), the difference was significantly greater in H5-vaccinated individuals than in H2-vaccinated individuals (Fig. 3f). Binding to H2 F45I stem was equivalent to H5 HA stem binding, again indicating that this residue is the key factor explaining reduced H2 stem binding in H5-vaccinated individuals (Fig. 3e,f). We noted overall lower HA stem-binding levels in protein-primed individuals than in DNA-primed individuals, but, in both vaccine prime groups, H2 and H5 HA stem binding were more similar in H2-vaccinated individuals than in H5-vaccinated individuals (Supplementary Fig. 2a–d). We conclude that vaccination with H2 leads to greater breadth in serological binding and neutralization than with H5 vaccination.
a,b, Sera were tested for binding to full-length H2 or H5 HA. Shown is the area under the curve (AUC) of serological binding as determined by MSD (a) or the log2 ratio of AUC between H5 and H2 binding (b) in each individual plotted separately for each vaccine group. c,d, Sera were tested for the capacity to neutralize H2 or H5 virus in a microneutralization assay. Shown is the 80% inhibitory concentration (IC80) (c) or ratio of H2 to H5 IC80 (d) in each vaccine group. e,f, Sera were tested for binding to H5, H2 WT, H2 F45I or H1 HA stem. Shown is the AUC of binding in each vaccine group (e) or the log2 ratio of binding as indicated (f). All sera tested were from 2 weeks after the H2 or H5 vaccine boost (n = 15 per vaccine group). All data are representative of two or three replicates. Bars in b, d and f represent the mean. Statistical significance was determined by the two-sided Wilcoxon matched-pairs rank test (a, c, e) or the two-sided Mann–Whitney test (b, d, f); n.s., not significant.
VH1-69 usage dominates the HA stem-binding immunoglobulin response
To investigate whether the immunoglobulins produced by HA stem-reactive memory B cells elicited by the two vaccines are genetically different, we performed single-cell sorting of HA stem-specific IgG+ memory B cells from peripheral blood mononuclear cells (PBMCs) collected 2 weeks after the vaccine boost from 12 H5-vaccinated individuals and 13 H2-vaccinated individuals and sequenced the paired heavy and light chains. This analysis included a mix of individuals primed with the DNA and protein vaccines (Supplementary Table 1). We first observed that VH1-69 usage equally dominated the immunoglobulin variable heavy chain (VH) repertoire, with over 50% of all HA stem-reactive memory B cell immunoglobulins expressing this VH gene in both trials (Fig. 4a), whether individuals were primed with a DNA vaccine or a protein vaccine (Supplementary Fig. 3a,b). We then proceeded with a more in-depth analysis of the HA stem-specific memory B cell immunoglobulin repertoire. With index sorting, we were able to assign HA specificity to each sorted B cell, allowing us to compare the immunoglobulin repertoires of triple-positive (H2H5H1) cells elicited by H2 (n = 638) and H5 (n = 973) vaccination, as well as double-positive (H5H1) cells (n = 451) induced by H5 vaccination. Few double-positive H2H1 or H2H5 memory B cells were detected after H2 vaccination, and these cells were thus excluded. We first calculated the average clone size of the HA stem-specific repertoire across all individuals and found it to be significantly lower after H5 vaccination (1.6 and 2.1 for the H5 and H2 vaccine trials, respectively), suggesting a more clonally diverse response (Supplementary Fig. 3c). We further plotted the generalized Hill diversity index (qD) over a range of diversity orders (q)22,23, which takes into account both clonal size and number of unique clonotypes. We found that HA stem-specific memory B cells after H5 vaccination were more clonally diverse than after H2 vaccination independently of specificity, even when we equalized the number of cells analyzed from each trial or limited analysis to only VH1-69 immunoglobulins (Fig. 4b and Supplementary Fig. 3d,e). The average VH mutation percentage, however, was not significantly different among HA stem-binding memory B cells isolated from each trial, whether or not they bound H2 (Fig. 4c). We also observed similar VH usage among cells of different specificity, with VH1-69 usage making up 50–60% of that for both H5H1H2 and H5H1 stem-reactive memory B cells (Fig. 4d and Supplementary Fig. 3f). Thus, although the proportion of HA stem-binding memory B cells with a VH1-69-encoded immunoglobulin did not differ between the two trials, the cross-reactivity of the VH1-69 immunoglobulins was different. Whereas 95% of the VH1-69 immunoglobulins expressed by memory B cells elicited by H2 vaccination were able to bind H1, H2 and H5, 30% of VH1-69 HA stem-specific memory B cells expanded by H5 vaccination were unable to bind H2 by flow cytometry (Fig. 4e).
a, VH gene usage for all single sorted HA stem-reactive B cells in each vaccine study as indicated (n = 1,426 from 12 H5-vaccinated individuals and 677 heavy chain sequences from 13 H2-vaccinated individuals). b, Hill diversity index (qD) over a range of diversity orders (q) for HA stem-specific B cells from the H2 vaccine trial (orange) or the H5 vaccine trial (blue), with the indicated specificity. The curves show the mean from 1,000 bootstraps, and the bands indicate 95% confidence intervals. c, Percent VH mutation of immunoglobulins from HA stem-binding B cells isolated after vaccination, with the indicated specificity. Each dot is the mean VH mutation percentage for all stem-binding immunoglobulin sequences isolated from a given individual, with the bar indicating the mean of each group. No statistically significant difference was observed in percent VH mutation between HA stem-binding immunoglobulins from the H2 versus H5 vaccine trial by two-sided Mann–Whitney test or between H5H1- and H5H2H1-specific cells isolated from the same individual after H5 vaccination by two-sided Wilcoxon matched-pairs rank test. d, VH gene usage for HA stem-specific B cells from the H2 vaccine trial (orange) or the H5 vaccine trial (blue), with the indicated specificity. e, Proportion of VH1-69-expressing HA stem-binding B cells detected after H2 or H5 vaccination, with triple or double reactivity as indicated. f, Twenty-one mAbs isolated from VH1-69 stem-binding B cells sorted from H5-vaccinated individuals were tested for binding by MSD (left, in blue) and neutralization (right, in orange) for group 1 HA subtypes as indicated. For neutralization, mAbs were tested for the capacity to neutralize virus strains at four concentrations as indicated and the color corresponds to the smallest concentration at which there was a 50% or greater reduction in viral titer. The names of mAbs from B cells detected as negative for H2 binding by flow cytometry are in blue. g, Binding AUC and neutralization potency of 21 mAbs isolated from VH1-69 H2 HA stem-binding B cells detected in PBMCs after H2 vaccination. Asterisks in f and g indicate mAbs used in structural analysis.
To confirm specificity determined by flow cytometry, we cloned and expressed 21 VH1-69 HA stem-binding mAbs isolated after H5 vaccination from seven different individuals. We set a median fluorescence intensity (MFI) of 1,000 as the cutoff between H2 HA-binding and non-H2-binding B cells by flow cytometry and expressed 11 mAbs above and 10 mAbs below this cutoff (Supplementary Fig. 4a). We confirmed with a Meso Scale Discovery (MSD) binding assay that the ten mAbs expressed by B cells lacking binding to H2 HA in flow cytometry had no to low binding to H2 HA (Fig. 4f, in blue, and Supplementary Fig. 4a,b). One additional mAb (44-2F08) isolated from a B cell detected as binding to H2 also showed no binding to H2. These non-H2-binding mAbs, however, were able to bind the H2 F45I HA mutant, and all but 44-2F08 had reduced or no binding to an H5 I45F HA mutant (Supplementary Fig. 4b). Non-H2-binding mAbs were largely broadly binding across other group 1 subtypes, including H5, H1, H6 and H9 (Fig. 4f). H2 stem-binding VH1-69 mAbs isolated from H2-vaccinated individuals were also generally broadly binding across group 1 subtypes tested (Fig. 4g and Supplementary Fig. 4c). Neutralization of H1N1 and H5N1 viruses was equivalent between H2-binding and non-H2-binding mAbs, but fewer non-H2-binding mAbs were able to neutralize H6N1 (P = 0.04, Fisher’s t-test) or H9N2 (P = 0.02, Fisher’s t-test) viruses (Fig. 4f,g). Overall, we conclude that a subset of VH1-69-encoded HA stem-binding immunoglobulins elicited by H5 vaccination cannot accommodate the Phe45HA2 residue in H2 HA, and this is associated with lower breadth of neutralization for group 1 subtypes H6 and H9.
Structure of HA stem recognition by VH1-69 antibodies
To understand the structural basis for differences in H2 binding among VH1-69 antibodies, we determined the structures for two of the mAbs described above with key VH1-69 class characteristics, including a CDRH2 Phe54, a CDRH3 Tyr98 and a kappa light chain, but with differing ability to bind and neutralize H2. We obtained the structure of the non-H2-binding 30-1D06 Fab in complex with H1 (A/NewCaledonia/20/1999, NC99) HA and H2-binding 310-1B11 Fab in complex with H2 (A/Canada/720/2005, CAN05) HA by single-particle cryo-electron microscopy (cryo-EM) (Supplementary Fig. 5 and Supplementary Table 2). We found that both mAbs displayed the canonical recognition of VH1-69-derived antibodies involving heavy chain residues 53 and 54 (Fig. 5a,b)24,25,26. Specifically, we observed that the critical Ile45HA2-NC99 or Phe45HA2-CAN05 in the HA stem region of the epitope was recognized by a hydrophobic patch on the Fab heavy chain comprising Met53 (30-1D06) or Ile53 (310-1B11) and Phe54 (Fig. 5c). However, the two antibodies showed a 9° shift in orientation relative to each other (Fig. 5d), resulting in a shift of the 310-1B11 heavy chain residue Ile53HC away from HA and, thereby, accommodating the larger bulk of Phe45HA2-CAN05 versus the smaller Ile45HA2-NC99 (Fig. 5e). In addition, in 30-1D06, the germline heavy chain residue Ile53 was mutated somatically to a larger and longer methionine residue, which is able to pack more snugly with Ile45HA2 on H5 or H1 HAs but clashes with Phe45 on H2 HAs (Fig. 5e). To verify the effect of Met53HC on 30-1D06 binding to H2, we created 30-1D06 variants with the wild-type Met53HC residue altered to alanine, isoleucine and valine. Reverting Met53HC to the germline Ile53HC led to a small decrease in affinity to H1 and H5 HAs but, most notably, allowed binding of 30-1D06 to H2 HA (Kd of 16 nM) (Fig. 5g and Supplementary Fig. 6a,b). This indicates that the Ile53MetHC somatic mutation is a critical factor in preventing 30-1D06 from binding H2 strains.
a, Cryo-EM density is shown for binding of 030-1D06 Fab (blue and cyan) with the NC99 H1 HA trimer (protomers in gray; particles 32/154, left) and 310-1B11 Fab (orange and yellow) with the CAN05 H2 HA trimer (protomers in gray; particles 115/114, right). Color-coding in a is used for b–f. b, Surface representations of the bound HAs showing the 5-Å footprint of the Fab-binding site colored as in a with an outline of the alternate Fab shown for comparison. c, Detailed view of the binding epitopes surrounding residue 45. d, Overlay of the two antibodies showing a 9° difference in the approach angles from residue 45. Purple and green inset boxes are shown in greater detail in e and f, respectively, which are rotated ~20° for clarity. e, Phe45HA2 clashes with Met53 of the 30-1D06 heavy chain but not Ile53 of 310-1B11. f, A hydrophobic pocket at the HA1/HA2 interface is displayed with the trimer surface colored by electrostatic charge. The two Fab fragments engage the pocket through somatic mutation Il28HC of 310-1B11 (orange) overlayed with somatic mutation Phe74HC of 30-1D06 (blue). The germline residues are labeled the color of the Fab, with the mutated residues labeled in red. g, Binding of 30-1D06 with WT Met53 and Ile53, Val53 and Ala53 variants to H2 WT, H2 F45I, H5 and H1 HA by MSD. Shown is the mean value ± s.d. of mAbs tested in duplicate. Data are representative of two independent experiments. ECL, electrochemiluminescence. h, Manhattan plot of sequence-binding association analysis using 1,042 H2-binding and 251 non-H2-binding sequences. Amino acids below an adjusted P value of 0.01 are colored. Adjusted P values were determined by Fisher’s exact test and adjusted for multiple tests with Holm’s method. i, Table showing residues with an adjusted P value below the 0.01 cutoff with odds ratio, residue position in the heavy chain sequence and whether the residue is a germline mutation (yes) or a somatic mutation (no). Odds ratio >1 (blue) indicates association with H2 non-binding; odds ratio <1 (orange) indicates association with H2 binding.
Some VH1-69-class antibodies capable of binding H2 HA, however, had a methionine at heavy chain residue 53 (Supplementary Fig. 7), indicating that this somatic mutation does not universally prevent accommodation of H2 Phe45HA2. To gain insight into other residue differences that might be associated with binding or not binding H2, we analyzed VH1-69-encoded heavy chain sequences obtained from 1,042 H2-binding B cells and 251 non-H2-binding B cells, as detected by flow cytometry. Sequence-feature association analysis (SeqFeatR) of these sequences provided detailed insight into amino acid position combinations significantly associated with H2 binding or non-binding (Fig. 5h,i). Several residues identified were present in 310-1B11 and 30-1D06, including Met53HC and two others (Ser74PheHC and Thr28IleHC), which fill a second hydrophobic pocket on HA with opposite association to H2 binding (Fig. 5f and Supplementary Fig. 6c). This hydrophobic pocket is also filled by Phe74HC on two published VH1-69 mAbs with limited ability to neutralize H2 (refs. 13,15). Other residues identified have also been shown to have prevalent somatic mutations associated with increased affinity to the HA stem27,28. Overall, these data reveal the capacity of VH1-69 germline residues Ile53 and Phe54 to accommodate either Ile45HA2 or Phe45HA2 on the HA stem. However, somatic mutations can permit H2 binding or lead to clashes with H2 Phe45HA2, as in the case of Met53HC on 30-1D06. This also shows that multiple factors can lead to loss of binding to H2 HA and such loss of binding is more likely a combinatorial effect than the result of a single residue change.
H2 exposure enhances cross-reactivity of the H5 vaccine response
All individuals included in our analysis thus far were born after 1970 and, thus, naive to H2. However, we wondered whether pre-exposure to H2N2 virus 50 years ago might change the response to H5 vaccination. To investigate this, we analyzed the response to H5 vaccination in all individuals born before 1966 (n = 11, after excluding one vaccine non-responder) and compared it to that in individuals born after 1973 (n = 17) in matched vaccine groups (Supplementary Table 1). Analysis of H2 binding in sera before H5 vaccination showed that individuals born before 1966 did, for the most part, have higher levels of serum antibodies to H2 HA than individuals born after 1973 (Fig. 6a). The two individuals with lower H2-binding levels were born in 1965 and likely had less exposure to H2 before it stopped circulating in 1968 (Fig. 6b).
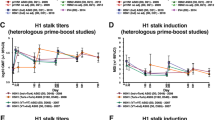
a,b, Sera collected before H5 vaccination in individuals born before 1966 or after 1973 were tested for reactivity to H2 HA or the H2 stem domain by MSD. Shown are the AUC of serological binding for the two age groups (a) and the AUC of H2 binding versus the year of birth (YOB) of all individuals (b). The vertical dotted line in b indicates the year H2N2 stopped circulating in the human population (1968). Data are representative of two or three independent experiments. c–e, Memory B cells from PBMCs collected 2 weeks after the H5 vaccine boost were tested by flow cytometry to measure the HA-specific response in the two age cohorts. Shown are the proportion of IgG+ B cells binding H5 HA (c) and the H5 stem domain (d) and the proportion of B cells binding the H5 stem domain with different specificities as indicated (e). f, Correlation between pre-vaccination H2 stem-binding serological levels and the proportion of HA stem-binding memory B cells that were H5, H2 and H1 cross-reactive 2 weeks after H5 vaccination. Statistical significance was determined using Spearman’s rank correlation coefficient. g,h, H5 stem-reactive B cells were isolated by single-cell sorting, and immunoglobulin genes were sequenced. Shown are the proportion of H5 stem-binding B cells in each individual that used VH1-69 (g) and the specificity as detected by flow cytometry of the VH1-69 H5 stem-binding B cells in the two age groups (h). i–l, Sera collected before vaccination and 2 weeks after boost were tested for binding levels to H5, H2 WT and H2 F45I stem proteins. Shown are the AUC of binding in each vaccine group (i,k) and the log2 ratio of H2 stem binding versus H5 stem binding (j,l). Data are representative of two independent experiments. Serum binding and B cell analysis in all panels was performed with the same individuals (n = 17 born after 1973 and n = 11 born before 1966). Statistical significance was determined by the two-sided Mann–Whitney test for a, c–e, j and l. The two-sided Wilcoxon matched-pairs rank test was used for i and k. Bars in a, c–e, g and i–l represent the mean.
We first compared the IgG+ memory B cell response to H5 vaccination in individuals from the two age cohorts 2 weeks after the H5 boost. The total H5 HA-specific and H5 HA stem-specific IgG+ memory B cell frequencies were quite variable, with no statistical difference between the two age groups (Fig. 6c,d). However, on average, 80% of H5 HA stem-specific memory B cells in individuals born before 1966 were cross-reactive with H1 and H2 (Fig. 6e), and the proportion of H5H1H2 cross-reactive memory B cells correlated with the pre-vaccination H2 stem serological binding levels (Fig. 6f). We did not detect a significant difference in VH1-69-expressing H5 HA stem-specific memory B cells between the two age cohorts (Fig. 6g), but, as in the case of H2 vaccine-elicited HA stem-specific memory B cells, 95% of VH1-69 stem-binding B cells from H5-vaccinated individuals born before 1966 were able to bind H5, H1 and H2 compared to 70% in individuals born after 1973 (Fig. 6h).
We also compared serological H2 HA stem-binding antibody levels between the two age cohorts in the H5 vaccine trial. Sera collected before H5 vaccination showed decreased H2 stem-binding levels in both age groups compared to H5 or H1 stem binding, but the reduction in H2 stem binding was significantly greater in the younger cohort naive to H2 (Fig. 6i,j). As H5 influenza virus has not circulated in the US population, H5 stem specificity detected before vaccination in either cohort would be primarily from cross-reactive antibodies generated by H1 exposure in younger individuals and H1 and/or H2 exposure in older individuals. Upon H5 vaccination, HA stem-binding antibody levels increased in both age cohorts (Fig. 6k), but the fold decrease in H2 stem-binding levels compared to H5 stem binding remained significantly greater in H2-naive individuals than in the older cohort (Fig. 6l). We propose that H5 vaccination is largely boosting pre-existing stem-directed memory B cells with differing levels of H2 cross-reactivity determined by influenza exposure early in life.
Discussion
Here we show that, by immunizing with H2, which naturally contains Phe45HA2, we can selectively expand memory B cells with greater breadth of reactivity and neutralization without compromising the magnitude of the total HA stem-specific memory B cell response. One difference between the H2 and H5 vaccine trials was the use of different protein immunogens. However, we think it is unlikely that this difference in the protein immunogen accounts for our results. We observed no difference in the overall magnitude or VH repertoire of the HA-specific responses, and the proportion of HA stem-binding memory B cells was very similar between the two trials.
We were somewhat surprised that the VH1-69 immunoglobulin class dominated the HA stem-specific memory B cell response after H2 vaccination, given the centrality of the bulky Phe45HA2 residue in the primary HA stem-binding epitope and multiple reports of otherwise broadly reactive HA stem-binding antibodies failing to recognize H2 (refs. 13,14,16). The cryo-EM structure of a VH1-69 antibody in complex with H2 HA indicated that the germline Ile53-Phe54 residues in the VH1-69 CDRH2 that make contact with the HA stem around Phe45HA2 can accommodate the bulky phenylalanine aromatic residue with a minor change in the Fab approach orientation. Other germline residues important for binding, such as Phe29HC, Phe54HC and Tyr98HC, were also not significantly different between H2-binding and non-H2-binding B cells. This suggests that initial engagement of VH1-69 germline HA stem-binding immunoglobulins with H2 HA is similar to that for H1 or H5 HA. However, somatic mutations that optimize the interface between an immunoglobulin and an HA with Ile45HA2 can lead to reduced binding to H2 HA.
The increased breadth that we observed with H2 vaccination extended to the entire HA stem-specific B cell response, not just the VH1-69 response. Addition of Phe45HA2 selects for memory B cells with immunoglobulins that have the most flexibility for accommodating residue changes, without compromising the magnitude or potency of the memory B cell response. Notably, the antibody response to an HA stem immunogen with Phe45HA2 might be more resistant to viral antibody escape mutants. H3 Ile45PheHA2 escape mutants have been identified in antibody resistance screens with the very broadly reactive antibodies CR9114 and FI6v3 (ref. 11), and we show that H5 Ile45PheHA2 abolishes binding of some HA stem-specific B cells. The emergence of H2N2 in the human population in 1957 might have been facilitated by the presence of Phe45HA2.
In 1960, Thomas Francis Jr. first proposed that the initial influenza virus strain(s) that a person is exposed to as a child establish an antibody repertoire that governs subsequent antibody responses to influenza strains throughout life. This concept has been validated in multiple studies29,30,31,32,33. One study looking at avian-origin H5N1 infections in Asia showed that childhood exposure to group 1 H1N1 or H2N2, but not group 2 H3N2, was associated with protection from serious group 1 H5N1 disease34. This group-level protection is postulated to be mediated, in part, by HA stem-specific antibodies established early in life. However, direct evidence that the HA stem-specific repertoire in adults is shaped by childhood influenza exposure has not been shown. Here we show distinct differences in the HA stem-specific response to H5 vaccination depending on childhood exposure to H2 or H1 influenza subtypes. In individuals exposed to H2 as children, despite exposure to H1 for many years subsequently, both the cellular and serological HA stem-specific responses to H5 vaccination had greater cross-reactivity to H2 than in individuals born when H1 and/or H3 were circulating in the human population.
Although we show that childhood exposure can establish a lifelong B cell repertoire compatible with Phe45HA2, it is unclear whether immunization of adults with H2 will also lead to the establishment of long-lasting memory B cells recognizing the H2 HA stem domain. Long-term follow-up of younger adults who participated in our H2 vaccine trial will be needed to understand whether immunization in adults can have lasting effects on the HA stem-binding B cell repertoire. All together, these data suggest a benefit from using H2 HA as an HA stem-based immunogen for eliciting the greatest breadth of reactivity across group 1 subtypes. If used in combination with a group 2 immunogen, this could lead to broad protection against influenza A.
Methods
Vaccine study design
The H5 vaccine study17,19 (NCT01086657) and the H2 vaccine study (NCT03186781) were conducted at the National Institutes of Health (NIH) Clinical Center by the Vaccine Research Center Clinical Trials Program of the National Institute of Allergy and Infectious Diseases (NIAID). The trial protocols were reviewed and approved by the NIAID institutional review board. Informed consent was obtained from every enrolled individual and complied with all relevant ethical regulations. Compensation was given for time and effort related to participation in the clinical trial. Both studies were phase 1, open-label, randomized clinical trials in healthy adults designed to study the safety, tolerability and immunogenicity of prime–boost vaccination regimens. In the H5N1 vaccine study, individuals were vaccinated with a recombinant DNA plasmid that encodes H5 A/Indonesia/05/2005 or a monovalent influenza subunit virion (MIV; A/Indonesia/05/2005) vaccine manufactured by Sanofi Pasteur. All individuals were then boosted 4–24 weeks later with the MIV vaccine. Only individuals with a prime–boost interval of 16 or 24 weeks were included in the comparison with the H2 vaccine study. In the H2 vaccine study, individuals were vaccinated with a recombinant DNA plasmid that encodes H2 A/Singapore/1/1957 or an H2 HA ferritin nanoparticle. All individuals were then boosted 16 weeks later with the H2 HA ferritin nanoparticle. Only individuals aged 18–47 (year of birth, 1970–1999) from this trial were included.
Flow cytometry and single-cell sorting
Cryopreserved PBMCs from blood collected 2 weeks after the boost from trial individuals were stained with anti-human monoclonal antibodies to CD3 (1:400 dilution), CD56 (1:200 dilution), CD14 (1:200 dilution) and CD20 (1:400 dilution) from BioLegend; IgG (1:100 dilution) and IgM (1:40 dilution) from BD Biosciences; and CD19 (1:50 dilution) from Beckman Coulter. HA probes were expressed, biotinylated and labeled with fluorochromes, as described previously10. Aqua dead cell stain was added for live/dead discrimination (Thermo Fisher Scientific). Stained samples were run on a FACSAria II (BD Biosciences) running BD FACSDiva software 8.0, and data were analyzed using FlowJo version 10 (TreeStar). CD3−CD14−CD56−CD19+CD20+IgG+IgM− memory B cells were gated, and HA-binding memory B cells were single-cell sorted into 96-well plates. Addition of multiple HA probes with different fluorochromes and the ability of the FACSAria to detect and record the MFI of each fluorochrome individually for each sorted cell (index sorting) were used to determine the binding of each sorted B cell to HAs of multiple subtypes simultaneously. The following soluble fluorochrome-labeled HAs were used: H2 A/Singapore/2/1957 ectodomain and stabilized stem; H2 A/Singapore/2/1957 F45I mutant ectodomain and stabilized stem; H5 A/Indonesia/05/2005 ectodomain and stabilized stem; H1 A/NewCaledonia/20/1999 ectodomain and stabilized stem; and H1 A/Michigan/45/2015 ectodomain.
Single-cell immunoglobulin amplification and sequencing and mAb production
Reverse transcription was performed on sorted cells, and multiplexed PCR was used to amplify immunoglobulin heavy and light chain genes, as described previously35,36. We obtained paired heavy and light chain immunoglobulin sequences from an average of 70% of the single cells on which we performed PCR. PCR products were sequenced by Beckman Coulter or GENEWIZ.
Heavy and light chain sequences were synthesized and cloned by GenScript into IgG1 kappa or lambda expression vectors. To produce mAbs recombinantly, Expi293 cells (Thermo Fisher Scientific, cat. no. A14527) were transfected with plasmids encoding immunoglobulin heavy and light chain pairs with ExpiFectamine (Thermo Fisher Scientific). Monoclonal antibodies were purified from the cell supernatant using Protein A Sepharose (Pierce).
Immunoglobulin sequence analysis
Heavy and light chain sequences from single-cell PCR were aligned to germline sequences and annotated using IMGT to determine gene usage and VH mutation frequency37. Clonally related sequences were determined using the Immcantation Change-O package, using the DefineClones function with a Hamming distance of 0.15 (ref. 38). To determine the clonal diversity, the ‘alphaDiversity’ function from Alakazam version 1.0.1 used q values 1–5 sampled at intervals of 0.1. To correct for varying sequencing depths between the vaccine and HA stem specificity groups, 1,000 bootstraps were performed to repeatedly and uniformly subsample all the sequences23.
HA binding assay
MSD 384-well streptavidin-coated SECTOR Imager 6000 reader plates were blocked with 5% MSD Blocker A for 30–60 min and then washed six times with the wash buffer (PBS + 0.05% Tween). The plates were then coated with biotinylated HA protein (same protein as was used for flow cytometry) for 1 h, and washed. mAbs were diluted in 1% MSD Blocker A to 1 μg ml−1, serially diluted threefold and added to the coated plates. Sera were diluted 1:100 in 1% MSD Blocker A and serially diluted threefold before addition to coated plates. After a 1-h incubation, plates were washed and incubated with SULFO-TAG-conjugated anti-human IgG for 1 h. After washing, the plates were read using 1× MSD Read Buffer using an MSD SECTOR Imager 600. Binding curves were plotted, and the AUC was determined using Prism 8. The same HA strains listed for flow cytometry were used for binding assays, with the addition of H6 A/Taiwan/1/2013 (TW13) ectodomain, H9 A/Guangxi-Xiangshan/11522/2018 (GX18) ectodomain and A/California/04/2009 (CA09) ectodomain.
Biolayer interferometry
Analyses of binding kinetics of HA to antibodies were carried out using a ForteBio Octet HTX instrument. Assays with agitation set at 1,000 r.p.m. in PBS were performed at 30 °C in solid black tilted-bottom 384-well plates (Geiger Bio-One). Histidine-tagged HA trimer protein (30 μg ml−1) was captured on NTA sensor tips (ForteBio) for 180 s to a final mean signal level of 0.8–1.2 nm. After loading, the sensor tips were incubated in PBS for 60 s for baseline adjustment. Then, the trimer-coated tips were dipped into eight concentrations (500 nM, 250 nM, 125 nM, 62.5 nM, 31.25 nM, 15.625 nM, 7.8125 nM and 0 nM) of IgG diluted in PBS for association for 300 s, followed by dissociation for 300 s in PBS. A 1:1 curve fitting model was used to determine the apparent Kd using Octet software version 12.0.
Microneutralization
Generation of replication-restricted reporter (R3) viruses was described previously39. In brief, the viral genomic RNA encoding functional HA (R3ΔHA viruses) or PB1 (R3ΔPB1 viruses) was replaced with a gene encoding a fluorescent protein and viruses were then rescued and propagated in cell lines stably expressing the corresponding HA (R3ΔHA viruses) or PB1 (R3ΔPB1 viruses) by the reverse genetics technique. Viral stocks were titered by determining the number of fluorescent foci. MDCK-SIAT1 cells (Millipore Sigma, cat. no. 05071502) expressing PB1 were seeded at a density of 10,000 cells per well in a 384-well black plate with a transparent bottom (Greiner) 24 h before infection and incubated at 37 °C in a 5% CO2 humidified atmosphere. Viruses were diluted using Opti-MEM supplemented with TPCK-treated trypsin (Sigma) and then added to pre-washed cell monolayer wells in quadruplicate. The final concentration of TPCK-treated trypsin was 1 μg ml−1. Plates were incubated for 18–22 h at 37 °C in a 5% CO2 humidified atmosphere. Fluorescent foci were then counted using a Celigo Image Cytometer (Nexcelom) with a customized red filter to detect mKate2/TdKatushka2 reporter signals. A/Singapore/1/1957 (H2N2 SI57) and A/Indonesia/05/2005 (H5N1 IN05) were R3ΔHA viruses, and A/California/04/2009 (H1N1 CA09), A/Taiwan/1/2013 (H6N1 TW13) and A/Guangxi-Xiangshan/11522/2018 (H9N2 GX18) were R3ΔPB1 viruses.
For microneutralization assays on sera, one part of serum samples was mixed with three parts of diluted receptor-destroying enzyme II (RDE II, Denka Seiken) and treated at 37 °C for 16–18 h, followed by heat inactivation at 56 °C for 40 min. RDE-treated sera were serially diluted in Opti-MEM and mixed 1:1 with R3ΔHA viruses (~80,000 fluorescent foci per milliliter) diluted in Opti-MEM supplemented with TPCK-treated trypsin (Sigma). The final concentration of TPCK-treated trypsin was 1 μg ml−1. After incubation at 37 °C in a 5% CO2 humidified atmosphere for 1 h, serum–virus mixtures were added to pre-washed cell monolayers in a 384-well plate as described above. Plates were incubated and counted as described above. The percent neutralization was calculated by constraining the virus control (virus + cells) as 0% and the cell control (cells only) as 100% and plotted against serum concentration. A curve fit was generated by a four-parameter nonlinear fit model in Prism (GraphPad). IC80 values were obtained from the curve fit for each serum sample.
To determine the neutralization capacity of the mAbs, each mAb was diluted in Opti-MEM to 20 μg ml−1, serially diluted and mixed 1:1 with viruses as described above. Three tenfold dilutions were made to test neutralization at 10 μg ml−1, 1 μg ml−1, 0.1 μg ml−1 and 0.01 μg ml−1. We report the concentration at which there was a 50% or greater reduction in virus levels compared to the no-antibody control cells.
Production of influenza HA proteins and Fab fragments for cryo-EM
DNA constructs encoding the HA ectodomains of H1 A/NewCaledonia/20/1999 (NC99) and H2 A/Canada/720/2005 (CAN05) with a C-terminal thrombin cleavage sequence, followed by a T4 fibritin trimerization motif and a hexa-histidine (6×His) affinity tag, were cloned into a pVRC8400 expression plasmid as previously described10. Two cysteine mutations were introduced at positions 30HA1 and 47HA2 in both the NC99 and CAN05 constructs to stabilize trimer formation by formation of an inter-protomer disulfide bond40. Additionally, a 6R furin cleavage site was added to enable efficient HA1 and HA2 processing. For expression, 700 mg L−1 of HA plasmid and 300 mg L−1 of furin construct were co-transfected into HEK293F cells using Turbo293 Transfection Reagent (Speed BioSystems) and cells were incubated at 37 °C for 6 d.
After cell culture collection, the supernatant was loaded onto cOmplete His-Tag Resin (Roche) and purified by gravity flow. HA protein was eluted from the column in PBS containing 300 mM imidazole and was later concentrated and further purified on a Superdex 200 16/60 size exclusion column (GE Healthcare).
To produce Fab fragments, the mAbs produced as described above were cleaved by LysC enzyme (1:4,000 wt/wt) (Roche) at 37 °C overnight. The enzymatic digestion was stopped by addition of protease inhibitor (Roche). The digestion mixture was then passed through a Protein A column to separate the Fc fragment from the Fab fragment. The Fab fragment was further purified with a Superdex 200 16/60 column in PBS.
Cryo-EM
Disulfide-stabilized HA trimers of strains H1 NC99 and H2 CAN05 were each incubated with twofold molar excess of Fab fragments of 030-1D06 and 310-1B11, respectively. Complexes were concentrated to 2 mg ml−1, and 2.3 µl of each was deposited on C-flat 1.2/1.3 grids (https://www.protochips.com/). Grids were vitrified in ethane with an FEI Vitrobot Mark IV plunger with a wait time of 30 s, blot time of 3 s and blot force of 1. Data collection was performed on a Titan Krios electron microscope with Leginon version 3.4 (ref. 41) using Gatan K2 Summit and Gatan K3 direct detection devices. Exposures were collected in movie mode for 10 s with the total dose of 70.13 e– per Å2 fractionated over 50 raw frames for the 030-1D06 complex and 2 s with a total dose of 51.16 e– per Å2 fractionated over 40 raw frames for the 310-1B11 complex. Images were pre-processed using Appion version 3.4 (refs. 42,43); frame alignments and dose weightings were performed using MotionCor2 (ref. 44). CTFFind4 (refs. 45,46) was used to estimate the initial CTF. cryoSPARC version 3.1 (ref. 47) was used for patch CTF estimation, particle picking, 2D classifications, ab initio 3D reconstruction, homogeneous refinement and nonuniform 3D refinement. Initial 3D reconstruction was performed using C1 symmetry, confirming three Fab molecules per trimer. Final refinements were performed using C3 symmetry.
Coordinates from PDB ID 3KU3 (ref. 48) were used for initial HA fit to the reconstructed maps. This was followed by simulated annealing and real-space refinement in Phenix version 1.19.2 (ref. 49) using the sharpened map from cryoSPARC version 3.1 and the density-modified map from Resolve in Phenix version 1.19.2 (ref. 50). The coordinates were then iteratively fit in Coot version 0.9 (ref. 51) and refined in Phenix version 1.19.2. Geometry and map fitting were evaluated throughout the process using Molprobity52 and EMRinger53 in Phenix version 1.19.2. PyMOL version 2.4.2 (www.pymol.org) and ChimeraX version 1.3 (ref. 54) were used to generate figures.
SeqFeatR
VH1-69-encoded immunoglobulin heavy chain sequences from single sorted B cells from individuals vaccinated with H2 or H5 were divided into H2-binding or non-H2-binding immunoglobulins as determined by flow cytometry. An in-house version of the R package SeqFeatR (https://seqfeatr.zmb.uni-due.de)55 was used to determine which heavy chain residues were associated with H2 binding and non-binding. In brief, for each residue position, we created a contingency table based on amino acid occurrence in H2-binding or non-H2-binding sequences and evaluated the association using Fisher’s exact test. The resulting P values were corrected using Holm’s multiple-testing method. The adjusted P values estimated for each amino acid position pair are represented in a Manhattan plot.
Statistical analysis
Statistical analysis was performed with Prism 8.0. Specific details of statistical analysis are indicated in the figure legends and the Results section. P values less than or equal to 0.05 were considered significant. Normality tests were conducted on all data to determine the appropriate statistical test. All statistical tests used were two tailed.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Cryo-EM maps and fitted coordinates were deposited in the Electron Microscopy Data Bank with accession codes 23098 and 23816 and in the Protein Data Bank with IDs 7L0L and 7MFG. Immunoglobulin sequences are available in GenBank under accession numbers OK669229–OK670607. Source data for Figs. 2–6 and Supplementary Figs. 1–4 and 6 are included as supplementary files. Any additional data are available upon reasonable request from the corresponding authors. Source data are provided with this paper.
References
Iuliano, A. D. et al. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet 391, 1285–1300 (2018).
Ellebedy, A. H. et al. Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proc. Natl Acad. Sci. USA 111, 13133–13138 (2014).
Li, G. M. et al. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc. Natl Acad. Sci. USA 109, 9047–9052 (2012).
Wrammert, J. et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J. Exp. Med. 208, 181–193 (2011).
Andrews, S. F. et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci. Transl. Med. 7, 316ra192 (2015).
Andrews, S. F. et al. Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans. Sci. Immunol. 2, eaan2676 (2017).
Avnir, Y. et al. IGHV1-69 polymorphism modulates anti-influenza antibody repertoires, correlates with IGHV utilization shifts and varies by ethnicity. Sci. Rep. 6, 20842 (2016).
Cortina-Ceballos, B. et al. Longitudinal analysis of the peripheral B cell repertoire reveals unique effects of immunization with a new influenza virus strain. Genome Med. 7, 124 (2015).
Wheatley, A. K. et al. H5N1 vaccine-elicited memory B cells are genetically constrained by the IGHV locus in the recognition of a neutralizing epitope in the hemagglutinin stem. J. Immunol. 195, 602–610 (2015).
Whittle, J. R. et al. Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J. Virol. 88, 4047–4057 (2014).
Wu, N. C. et al. Different genetic barriers for resistance to HA stem antibodies in influenza H3 and H1 viruses. Science 368, 1335–1340 (2020).
Corti, D. et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333, 850–856 (2011).
Dreyfus, C. et al. Highly conserved protective epitopes on influenza B viruses. Science 337, 1343–1348 (2012).
Joyce, M. G. et al. Vaccine-induced antibodies that neutralize group 1 and group 2 influenza A viruses. Cell 166, 609–623 (2016).
Lang, S. et al. Antibody 27F3 broadly targets influenza A group 1 and 2 hemagglutinins through a further variation in VH1-69 antibody orientation on the HA stem. Cell Rep. 20, 2935–2943 (2017).
Throsby, M. et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE 3, e3942 (2008).
Ledgerwood, J. E. et al. DNA priming and influenza vaccine immunogenicity: two phase 1 open label randomised clinical trials. Lancet Infect. Dis. 11, 916–924 (2011).
Wei, C. J. et al. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science 329, 1060–1064 (2010).
Ledgerwood, J. E. et al. Prime-boost interval matters: a randomized phase 1 study to identify the minimum interval necessary to observe the H5 DNA influenza vaccine priming effect. J. Infect. Dis. 208, 418–422 (2013).
Yassine, H. M. et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 21, 1065–1070 (2015).
Corbett, K. S. et al. Design of nanoparticulate group 2 influenza virus hemagglutinin stem antigens that activate unmutated ancestor B cell receptors of broadly neutralizing antibody lineages. MBio 10, e02810–18 (2019).
Hill, M. O. Diversity and evenness: a unifying notation and its consequences. Ecology 54, 427–432 (1973).
Stern, J. N. et al. B cells populating the multiple sclerosis brain mature in the draining cervical lymph nodes. Sci. Transl. Med. 6, 248ra107 (2014).
Ekiert, D. C. et al. Antibody recognition of a highly conserved influenza virus epitope. Science 324, 246–251 (2009).
Lingwood, D. et al. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature 489, 566–570 (2012).
Sui, J. et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 16, 265–273 (2009).
Avnir, Y. et al. Molecular signatures of hemagglutinin stem-directed heterosubtypic human neutralizing antibodies against influenza A viruses. PLoS Pathog. 10, e1004103 (2014).
Pappas, L. et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature 516, 418–422 (2014).
Lessler, J. et al. Evidence for antigenic seniority in influenza A (H3N2) antibody responses in southern China. PLoS Pathog. 8, e1002802 (2012).
Li, Y. et al. Immune history shapes specificity of pandemic H1N1 influenza antibody responses. J. Exp. Med. 210, 1493–1500 (2013).
Miller, M. S. et al. Neutralizing antibodies against previously encountered influenza virus strains increase over time: a longitudinal analysis. Sci. Transl. Med. 5, 198ra107 (2013).
Carter, D. M. et al. Sequential seasonal H1N1 influenza virus infections protect ferrets against novel 2009 H1N1 influenza virus. J. Virol. 87, 1400–1410 (2013).
Linderman, S. L. & Hensley, S. E. Antibodies with ‘original antigenic sin’ properties are valuable components of secondary immune responses to influenza viruses. PLoS Pathog. 12, e1005806 (2016).
Gostic, K. M., Ambrose, M., Worobey, M. & Lloyd-Smith, J. O. Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science 354, 722–726 (2016).
Tiller, T. et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT–PCR and expression vector cloning. J. Immunol. Methods 329, 112–124 (2008).
Doria-Rose, N. A. et al. New member of the V1V2-directed CAP256-VRC26 lineage that shows increased breadth and exceptional potency. J. Virol. 90, 76–91 (2016).
Brochet, X., Lefranc, M. P. & Giudicelli, V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 36, W503–W508 (2008).
Gupta, N. T. et al. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics 31, 3356–3358 (2015).
Creanga, A. et al. A comprehensive influenza reporter virus panel for high-throughput deep profiling of neutralizing antibodies. Nat. Commun. 12, 1722 (2021).
Lee, P. S., Zhu, X., Yu, W. & Wilson, I. A. Design and structure of an engineered disulfide-stabilized influenza virus hemagglutinin trimer. J. Virol. 89, 7417–7420 (2015).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
Voss, N. R., Yoshioka, C. K., Radermacher, M., Potter, C. S. & Carragher, B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopyand TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166, 205–213 (2009).
Lander, G. C. et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102 (2009).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Xu, R., McBride, R., Paulson, J. C., Basler, C. F. & Wilson, I. A. Structure, receptor binding, and antigenicity of influenza virus hemagglutinins from the 1957 H2N2 pandemic. J. Virol. 84, 1715–1721 (2010).
Adams, P. D. et al. Recent developments in the PHENIX software for automated crystallographic structure determination. J. Synchrotron Radiat. 11, 53–55 (2004).
Terwilliger, T. C., Ludtke, S. J., Read, R. J., Adams, P. D. & Afonine, P. V. Improvement of cryo-EM maps by density modification. Nat. Methods 17, 923–927 (2020).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Davis, I. W., Murray, L. W., Richardson, J. S. & Richardson, D. C. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32, W615–W619 (2004).
Barad, B. A. et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Budeus, B., Timm, J. & Hoffmann, D. SeqFeatR for the Discovery of Feature-Sequence Associations. PLoS ONE 11, e0146409 (2016).
Acknowledgements
We thank D. Ambrozak for help with flow cytometry sorting. Support for this work was provided by the Intramural Research Programs of the Vaccine Research Center and the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. Some of this work was performed at the Simons Electron Microscopy Center and the National Resource for Automated Molecular Microscopy, located at the New York Structural Biology Center, supported by grants from the Simons Foundation (SF349247) and the National Institute of General Medical Sciences (GM103310), with additional support from NYSTAR and the New York State Assembly Majority. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization, S.F.A.; methodology, S.F.A. and A.C.; formal analysis, S.F.A., R.R., C.-H.S. and J.C.B.; investigation, S.F.A., J.E.R., J.G., R.A.G., C.S., F.C., L.Y.C., A.C., D.R.H., A.S.O., A.N. and T.Z.; resources, K.V.H., G.L.C. and J.E.L.; writing—original draft, S.F.A.; writing—review and editing, S.F.A., J.C.B., K.V.H., M.K., J.R.M., P.D.K. and A.B.M.; visualization, S.F.A. and J.G.; supervision, S.F.A., G.L.C., J.R.M., B.S.G., M.K., J.E.L., P.D.K. and A.B.M.; funding acquisition, A.B.M.
Corresponding authors
Ethics declarations
Competing interests
J.C.B., B.S.G., J.R.M., M.K. and P.D.K. are named inventors of US patents 9,441,019, 10,137,190 and 10,363,301 on influenza hemagglutinin nanoparticle vaccines and stabilized hemagglutinin stem trimers and of several pending applications on related technologies filed by the Department of Health and Human Services (National Institutes of Health).
Peer review information Nature Medicine thanks the anonymous reviewers for their contribution to the peer review of this work. Alison Farrell was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–7 and Supplementary Tables 1 and 2.
Supplementary Table 1
Statistical source data for Supplementary Fig. 1.
Supplementary Table 2
Statistical source data for Supplementary Fig. 2.
Supplementary Table 3
Statistical source data for Supplementary Fig. 3.
Supplementary Table 4
Statistical source data for Supplementary Fig. 4.
Supplementary Table 5
Statistical source data for Supplementary Fig. 6.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Rights and permissions
About this article
Cite this article
Andrews, S.F., Raab, J.E., Gorman, J. et al. A single residue in influenza virus H2 hemagglutinin enhances the breadth of the B cell response elicited by H2 vaccination. Nat Med 28, 373–382 (2022). https://doi.org/10.1038/s41591-021-01636-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-021-01636-8
This article is cited by
-
Vaccine design via antigen reorientation
Nature Chemical Biology (2024)
-
Antibodies to combat viral infections: development strategies and progress
Nature Reviews Drug Discovery (2022)
-
CD4 T cell epitope abundance in ferritin core potentiates responses to hemagglutinin nanoparticle vaccines
npj Vaccines (2022)
-
Exploring the landscape of immune responses to influenza infection and vaccination
Nature Medicine (2022)