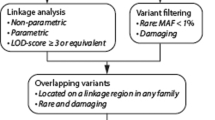
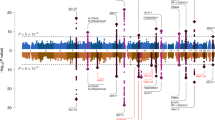
Abstract
Intracranial aneurysm (IA) rupture leads to subarachnoid hemorrhage, a sudden-onset disease that often causes death or severe disability. Although genome-wide association studies have identified common genetic variants that increase IA risk moderately, the contribution of variants with large effect remains poorly defined. Using whole-exome sequencing, we identified significant enrichment of rare, deleterious mutations in PPIL4, encoding peptidyl-prolyl cis-trans isomerase-like 4, in both familial and index IA cases. Ppil4 depletion in vertebrate models causes intracerebral hemorrhage, defects in cerebrovascular morphology and impaired Wnt signaling. Wild-type, but not IA-mutant, PPIL4 potentiates Wnt signaling by binding JMJD6, a known angiogenesis regulator and Wnt activator. These findings identify a novel PPIL4-dependent Wnt signaling mechanism involved in brain-specific angiogenesis and maintenance of cerebrovascular integrity and implicate PPIL4 gene mutations in the pathogenesis of IA.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All summary statistics for case–control burden analysis are fully detailed in Supplementary Tables 1–4. Each variant in Supplementary Tables 1 and 2 is annotated with information regarding genomic location, variant effect, amino acid change, CADD (version 3) score for deleteriousness and general and subpopulation MAF in gnomAD and ExAC. In addition, individual-level phenotypic data from patients with PPIL4 mutations are provided in Supplementary Table 3. Case–control burden analysis (two-tailed Fisher’s exact test) result for 17 genes that are co-segregating in all affected individuals with general gnomAD MAF < 0.005 is shown in Supplementary Table 3. Additionally, sequencing data of all patients in the IA cohort with PPIL4 mutations have been deposited in the European Genome-phenome Archive under accession number EGAS00001005518. Zebrafish expression data are reported in Supplementary Tables 5 and 6. Additionally, public expression and genomics datasets were obtained from the GtEX portal (https://gtexportal.org/home/), the database of gene expression in adult mouse brain and lung vascular and perivascular cell (https://betsholtzlab.org/VascularSingleCells/database.html) and the Genome Aggregation Database (https://gnomad.broadinstitute.org/). Gene enrichment analysis was performed using the Molecular Signatures Database (https://www.gsea-msigdb.org/gsea/msigdb/) and Metascape (https://metascape.org/gp/index.html#/main/step1). CRISPRscan was used for sgRNA design (https://www.crisprscan.org/). The BioPlex (version 3.0)(https://bioplex.hms.harvard.edu/explorer/home) database was used to explore potential protein interactors of PPIL4. VarCards67 (http://159.226.67.237/sun/varcards/) was used for variant annotation of the data individually downloaded from the gnomAD website. Source data are provided with this paper.
Code availability
In-house codes are available at https://doi.org/10.5281/zenodo.5539900.
References
Vlak, M. H., Algra, A., Brandenburg, R. & Rinkel, G. J. Prevalence of unruptured intracranial aneurysms, with emphasis on sex, age, comorbidity, country, and time period: a systematic review and meta-analysis. Lancet Neurol. 10, 626–636 (2011).
Korja, M., Lehto, H., Juvela, S. & Kaprio, J. Incidence of subarachnoid hemorrhage is decreasing together with decreasing smoking rates. Neurology 87, 1118–1123 (2016).
Lindbohm, J. V., Kaprio, J., Jousilahti, P., Salomaa, V. & Korja, M. Risk factors of sudden death from subarachnoid hemorrhage. Stroke 48, 2399–2404 (2017).
Korja, M. et al. Cause-specific mortality of 1-year survivors of subarachnoid hemorrhage. Neurology 80, 481–486 (2013).
Kissela, B. M. et al. Subarachnoid hemorrhage: a preventable disease with a heritable component. Stroke 33, 1321–1326 (2002).
Graf, C. J. Familial intracranial aneurysms: report of four cases. J. Neurosurg. 25, 304–308 (1966).
Schievink, W. I., Schaid, D. J., Michels, V. V. & Piepgras, D. G. Familial aneurysmal subarachnoid hemorrhage: a community-based study. J. Neurosurg. 83, 426–429 (1995).
Bor, A. S. E., Rinkel, G. J., van Norden, J. & Wermer, M. J. Long-term, serial screening for intracranial aneurysms in individuals with a family history of aneurysmal subarachnoid haemorrhage: a cohort study. Lancet Neurol. 13, 385–392 (2014).
Santiago-Sim, T. et al. THSD1 (thrombospondin type 1 domain containing protein 1) mutation in the pathogenesis of intracranial aneurysm and subarachnoid hemorrhage. Stroke 47, 3005–3013 (2016).
Bourcier, R. et al. Rare coding variants in ANGPTL6 are associated with familial forms of intracranial aneurysm. Am. J. Hum. Genet. 102, 133–141 (2018).
Zhou, S. et al. RNF213 is associated with intracranial aneurysms in the French-Canadian population. Am. J. Hum. Genet. 99, 1072–1085 (2016).
Yasuno, K. et al. Genome-wide association study of intracranial aneurysm identifies three new risk loci. Nat. Genet. 42, 420 (2010).
Yasuno, K. et al. Common variant near the endothelin receptor type A (EDNRA) gene is associated with intracranial aneurysm risk. Proc. Natl Acad. Sci. USA 108, 19707–19712 (2011).
Bilguvar, K. et al. Susceptibility loci for intracranial aneurysm in European and Japanese populations. Nat. Genet. 40, 1472 (2008).
McKenna, A. et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491 (2011).
Van der Auwera, G. A. et al. From FastQ data to high‐confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43, 11.10. 11–11.10. 33 (2013).
Krumm, N. et al. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 47, 582 (2015).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Cras, T. Y. et al. Determinants of the presence and size of intracranial aneurysms in the general population: the Rotterdam Study. Stroke 51, 2103–2110 (2020).
Taylor, C. L., Yuan, Z., Selman, W. R., Ratcheson, R. A. & Rimm, A. A. Cerebral arterial aneurysm formation and rupture in 20,767 elderly patients: hypertension and other risk factors. J. Neurosurg. 83, 812–819 (1995).
Zeng, L. et al. Molecular cloning, structure and expression of a novel nuclear RNA-binding cyclophilin-like gene (PPIL4) from human fetal brain. Cytogenetic Genome Res. 95, 43–47 (2001).
Hanes, S. D. Prolyl isomerases in gene transcription. Biochim. Biophys. Acta 1850, 2017–2034 (2015).
Lang, K., Schmid, F. X. & Fischer, G. Catalysis of protein folding by prolyl isomerase. Nature 329, 268–270 (1987).
Ando, K. et al. Clarification of mural cell coverage of vascular endothelial cells by live imaging of zebrafish. Development 143, 1328–1339 (2016).
Zhang, Y. et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014).
Vanlandewijck, M. et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480 (2018).
He, L. et al. Single-cell RNA sequencing of mouse brain and lung vascular and vessel-associated cell types. Sci. Data 5, 180160 (2018).
DeCicco-Skinner, K. L. et al. Endothelial cell tube formation assay for the in vitro study of angiogenesis. J. Vis. Exp. e51312 (2014).
Hillen, B., Drinkenburg, B. A., Hoogstraten, H. W. & Post, L. Analysis of flow and vascular resistance in a model of the cricle of Willis. J. Biomech. 21, 807–814 (1988).
Kamoun, W. S. et al. Simultaneous measurement of RBC velocity, flux, hematocrit and shear rate in vascular networks. Nat. Methods 7, 655 (2010).
Van Dijk, D. et al. Recovering gene interactions from single-cell data using data diffusion. Cell 174, 716–729 (2018).
Liberzon, A. In: Stem Cell Transcriptional Networks 153–160 (Springer, 2014).
Daneman, R. et al. Wnt/β-catenin signaling is required for CNS, but not non-CNS, angiogenesis. Proc. Natl Acad. Sci. USA 106, 641–646 (2009).
Stenman, J. M. et al. Canonical Wnt signaling regulates organ-specific assembly and differentiation of CNS vasculature. Science 322, 1247–1250 (2008).
Boeckel, J.-N. et al. Jumonji domain-containing protein 6 (Jmjd6) is required for angiogenic sprouting and regulates splicing of VEGF-receptor 1. Proc. Natl Acad. Sci. USA 108, 3276–3281 (2011).
Schneider, J. E. et al. Identification of cardiac malformations in mice lacking Ptdsr using a novel high-throughput magnetic resonance imaging technique. BMC Dev. Biol. 4, 16 (2004).
Zhou, D. et al. Inhibition of JMJD6 expression reduces the proliferation, migration and invasion of neuroglioma stem cells. Neoplasma 64, 700–708 (2017).
Zhang, X. et al. JmjC domain-containing protein 6 (Jmjd6) derepresses the transcriptional repressor transcription factor 7-like 1 (Tcf7l1) and is required for body axis patterning during Xenopus embryogenesis. J. Biol. Chem. 290, 20273–20283 (2015).
Zhang, Z., Yang, Y. & Zhang, X. MiR-770 inhibits tumorigenesis and EMT by targeting JMJD6 and regulating WNT/β-catenin pathway in non-small cell lung cancer. Life Sci. 188, 163–171 (2017).
Huttlin, E. L. et al. Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505 (2017).
Huttlin, E. L. et al. The BioPlex network: a systematic exploration of the human interactome. Cell 162, 425–440 (2015).
Böse, J. et al. The phosphatidylserine receptor has essential functions during embryogenesis but not in apoptotic cell removal. J. Biol. 3, 15 (2004).
Akhtar, S., Gremse, F., Kiessling, F., Weber, C. & Schober, A. CXCL12 promotes the stabilization of atherosclerotic lesions mediated by smooth muscle progenitor cells in Apoe-deficient mice. Arterioscler. Thromb. Vasc. Biol. 33, 679–686 (2013).
Vanhollebeke, B. et al. Tip cell-specific requirement for an atypical Gpr124-and Reck-dependent Wnt/β-catenin pathway during brain angiogenesis. eLife 4, e06489 (2015).
Cho, C., Smallwood, P. M. & Nathans, J. Reck and Gpr124 are essential receptor cofactors for Wnt7a/Wnt7b-specific signaling in mammalian CNS angiogenesis and blood–brain barrier regulation. Neuron 95, 1056–1073 (2017).
Moro, E. et al. In vivo Wnt signaling tracing through a transgenic biosensor fish reveals novel activity domains. Dev. Biol. 366, 327–340 (2012).
Sato, N., Meijer, L., Skaltsounis, L., Greengard, P. & Brivanlou, A. H. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 10, 55–63 (2004).
Kircher, M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310 (2014).
Landrum, M. J. et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46, D1062–D1067 (2017).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904 (2006).
Wang, C. et al. Ancestry estimation and control of population stratification for sequence-based association studies. Nat. Genet. 46, 409 (2014).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Chen, X. et al. MLL-AF9 initiates transformation from fast-proliferating myeloid progenitors. Nat. Commun. 10, 1–15 (2019).
Mootha, V. K. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Moon, K. R. et al. Visualizing structure and transitions in high-dimensional biological data. Nat. Biotechnol. 37, 1482–1492 (2019).
Moreno-Mateos, M. A. et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR–Cas9 targeting in vivo. Nat. Methods 12, 982 (2015).
Narayanan, A. et al. In vivo mutagenesis of miRNA gene families using a scalable multiplexed CRISPR/Cas9 nuclease system. Sci. Rep. 6, 32386 (2016).
Kasper, D. M. et al. MicroRNAs establish uniform traits during the architecture of vertebrate embryos. Dev. Cell 40, 552–565. e555 (2017).
Brend, T. & Holley, S. A. Zebrafish whole mount high-resolution double fluorescent in situ hybridization. J. Vis. Exp. 1229 (2009).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21 (2014).
Smedley, D. et al. BioMart—biological queries made easy. BMC Genomics 10, 1–12 (2009).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1–10 (2019).
Li, J. et al. VarCards: an integrated genetic and clinical database for coding variants in the human genome. Nucleic Acids Res. 46, D1039–D1048 (2018).
Acknowledgements
We are indebted to the patients and families who have contributed to this study. We would like to thank M. Cavanaugh for the zebrafish husbandry. Funding: This work was supported by Yale University funds (to M. Günel) and National Institutes of Health grants 4R01NS057756-10 (to M. Günel.), 1R01NS111935-01 (to M. Günel, A.L. and K.M.-G.) and R01 NS109160-04 and R01 HL130246-05 (to S.N.).
Author information
Authors and Affiliations
Contributions
T.B., M. Günel and S.N. conceptualized the study. T.B. performed genetic analysis and identified PPIL4 mutations; designed, performed and analyzed in vitro and in vivo experiments and data visualization; analyzed statistical data; and wrote the manuscript and the revision. E.R. and A.G.E.-S helped generate the ppil4 mutant line, performed in vivo experiments (zebrafish) and performed data visualization and analysis. D.F.M. analyzed bulk and scRNA-seq data and performed data visualization. C.N.-W., W.D. and S.C.J. analyzed genetic data. A.P. helped generate the jmjd6 mutant line and helped with data visualization. W.A. conducted the experiments in adult zebrafish. O.H. generated and validated all constructs used in the study. E.Z.E.-O. and A.S.H. analyzed RNA-seq. M. Guy, B. Gültekin, D.K., D.K.R., N.G., S.M.A., B. Gülez, S.A., K.O., Y.Y., S.C., E.S., E.D. and J.H. assisted with experimental work. A.C edited the manuscript. A.K.O. assisted with experimental work. A.L. edited the manuscript and the revision and supervised experimental work. K.B. and K.T.K. edited the manuscript and the revision and supervised experimental work. E.S.C.J. diagnosed patients and provided radiologic and clinical information. R.P.L. and M.K.K. supervised the research. K.Y. performed genetic analysis. K.M.G identified and resolved cell signaling mechanisms and designed and performed the signaling, localization and protein–protein interaction experiments. S.N. designed, analyzed, conducted and supervised zebrafish experiments, wrote and reviewed the manuscript and edited the revision. M. Günel recruited and clinically and radiologically evaluated patients with IA; analyzed the genetic data; supervised, wrote and reviewed the manuscript and the revision; and led the research.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Medicine thanks John Kolega, R. Loch Macdonald, Alan Shuldiner and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Anna Maria Ranzoni was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Sanger sequencing confirmation of PPIL4 variants identified by whole-exome sequencing (WES).
Sanger sequencing confirming heterozygous PPIL4 variants identified in IA patients. Locations of single nucleotide variations (red arrows), deletions (“:”) and the consecutive overlapping sequence. * represents stop gain mutation.
Extended Data Fig. 2 Gross morphologic assessment of ppil4 mutant zebrafish.
a, Schematic of the zebrafish ppil4 protein showing the location of the stop codon generated in the PPIase domain by Crispr-CAS9. A deletion of 11 bp in exon 5 causes a frameshift and a premature stop codon. b, Fluorescent in situ hybridization for ppil4 in ~1.3 and ~2 dpf wild type embryos (upper row). ppil4 is expressed in the head region but not in the trunk. ppil4 expression is lost in ppil4−/− mutants (lower row). The midbrain (blue-dashed line), hindbrain (pink-dashed), and optic tectum (white-dashed) boundaries are indicated, n= 3 sets of biological replicates with 30 zebrafish (each set) for both timepoints. c, Reduction in ppil4 expression in heterozygous and homozygous mutant zebrafish validated by qPCR. Values shown as fold change in different genotypes (X-axis) relative to wild type (Y-axis), beta actin was used for normalization, n= 3 sets of biological replicates. d, Bright-field images of wild-type, ppil4+/−, and ppil4−/− embryos at different stages of development. No gross morphological defects were observed in heterozygous mutants. Arrows (6 dpf): swim bladder. e, ppil4−/− zebrafish manifest necrosis in the head at 48 hpf (white arrow), not evident at 32 hpf and lower jaw abnormality at 6 dpf (white arrow), n=3 sets of biological replicates. Data presented as individual scatter plot with median. Statistical test: One-way ANOVA with Dunnett`s multiple comparison test. E= eye, OV= otic vesicle, OT= Optic tectum. Scale bar: 200 μm in b and e; 1 mm in d.
Extended Data Fig. 3 The impact of ppil4 depletion on cerebrovascular network is persistent at 5.5 dpf.
a, Left panels: Maximum intensity projection (MIP) of representative confocal z-stack images in 5.5 dpf old ppil4+/+ (n=6), ppil4+/− (n=5) and ppil4−/− (n=6) embryos in the tg(kdrl:gfp)zn1 background (dorsal-view and caudal facing left). Right panels: Brain vessel segmentation in the larvae shown at left (Imaris). Colors represent branch depth ranging from 0 to 12 (higher and lower branch depth shown in red and blue, respectively). b-e, Quantification of midbrain CtA branch number (b) and length (c); Quantification of hindbrain CtA branch number (d) and length (e) in ppil4+/+ (n=6), ppil4+/− (n=5) and ppil4−/− (n=6) embryos. f-h, Confocal images (f) and comparative assessment (g,h) of trunk vasculature in 5.5 dpf old ppil4+/+ (n=7), ppil4+/− (n=4) or ppil4−/− (n=4) zebrafish. i-k, Confocal images (i) and comparative assessment (j,k) of trunk vasculature in 2.5 dpf old, n=9 per genotype. Individual values shown with scatter dot plot and median in b-e. The box extends from the 25th to 75th percentile. The whiskers show the minimum and the maximum values, while the line in the middle of the box is median in g,h,j and k. Statistical tests: One-way ANOVA followed by Dunnett’s multiple correction for all comparisons. Scale bar: 50 μm in a and i, 100 μm in f.
Extended Data Fig. 4 Loss of ppil4 results in reduction in endothelial cell number.
a,b, Confocal images of the cerebral vasculature of (a) 30 and (b) 60 hpf zebrafish in the tg(kdrl:mCherry; fli1:nGFP) background, expressing mCherry and GFP respectively in the cell membrane and nuclei of endothelial cells (ECs) (dorsal view). Arrows indicate angiogenic sprouting in midbrain (a), and hindbrain CtAs (b). c, Comparison of EC number at 30 hpf ppil4+/+ (n=10), ppil4+/− (n=15), and ppil4−/− (n=3) embryos. d, Comparison of EC number in the cerebral arteries of 60 hpf ppil4+/+ (n=9), ppil4+/− (n=17), and ppil4−/− (n=7) embryos. Individual values presented with scatter dot plot and median for all quantifications. Statistical tests: One-way ANOVA followed by Dunnett’s multiple comparison test for all comparisons. Abbreviations: Mb= Midbrain, Hb= Hindbrain, BA= Basilar Artery, PCS= Posterior communicating segment, PMBC= Primordial midbrain channel, PHBC= Primordial hindbrain channel, CtA= Central Artery. Scale bar: 50 μm in a and b.
Extended Data Fig. 5 ppil4−/− zebrafish exhibit apoptosis in neurons and radial glia, but not in endothelial cells.
a,b, Representative confocal images of 2.5 dpf embryos, where ppil4−/− mutants exhibit an increase in TUNEL-positive cells in the head region, n=4 sets biological replicates with 30 zebrafish per set. c-j, Cross-sections of the head. Embryos at 2.5 dpf in tg(kdrl:gfp)zn1 background were stained for Caspase-3 and HU (neurons) or GFAP (radial glia). Apoptosis was detected in neurons and radial glia (white asterisks), but not in endothelial cells (white box; arrows). Confocal images, n= 3 sets biological replicates with 30 zebrafish per set. k,l, Whole mount confocal images of 2.5 dpf embryos showing no difference in (k) neuronal (HU+) and (l) radial glial (GFAP+) population in ppil4 mutant genotypes, n= 3 sets biological replicates with 30 zebrafish per set. Dorsal view of the head region (left panel), lateral view of the trunk (right panel). Individual values shown with scatter dot plot and median. Statistical test: (b) One-way ANOVA followed by Dunnett`s multiple comparison test. Abbreviations: TUNEL: Terminal deoxynucleotidyl transferase dUTP nick end labeling; Fb: forebrain; Mb: midbrain; Hb: hindbrain; e=eye. Scale bar: 50 μm.
Extended Data Fig. 6 PPIL4 expression in endothelial cells.
a, Percentile rank of 15,521 transcripts expressed (FPKM) in brain-enriched endothelial cells (EC) in wildtype zebrafish (2.5 dpf), n=4 sets of biological replicates. b, ppil4 expression in zebrafish brain-enriched EC’s at 12, 24, 72 hpf and 3 months (relative to beta actin levels), n=3 sets of biological replicates. c, ppil4 expression levels in kdrl-GFP+ endothelial cells in the brain, liver, and the heart in 3 months old tg(kdrl:gfp) zebrafish (relative to beta actin levels), n=3 sets of biological replicates. d-i, Double immunostaining with pan-endothelial marker CD31 and PPIL4 demonstrating overlap in endothelial cell layer of human middle cerebral artery, n=4 biological replicates. j,k,m, In vitro tube formation assay showing significant impairment in branch formation in shRNA-treated compared with non-target control shRNA treated HUVECs, n=5 biological replicates. l, Reduction in PPIL4 expression in shRNA treated HUVEC validated by qPCR. Values shown as fold change relative to control (relative to TATA-binding protein [TBP]); n=4 biological replicates. m, Reduced number of nodes, branches, and junctions upon PPIL4 downregulation; n=5 biological replicates. n, Expression of PPIL4 and a list of angiogenesis-associated genes in wild type HUVECs (relative to TBP); n= 3 biological replicates. Individual values shown with scatter plot as mean with standard deviation in b; and median in c,l,n. In m, the box extends between 25th-75th percentile. The whiskers show the minimum and the maximum values. Central line is median. Statistical tests: (b,c) One-way ANOVA followed by Dunnett’s multiple comparison. (l,m) Two-tailed Student t-test.
Extended Data Fig. 7 Endothelial cell-specific overexpression of PPIL4 in ppil4−/− embryos restores cerebrovascular network simplification.
a-e``` Maximum intensity projection (MIP) of five representative confocal z-stack images of UAS:hPPIL4-WT-tagRFP injected embryos in (ppil4−/- ; Tg(kdrl:gfp)zn1; tg(fli1a:gal4)) background at 2.5 dpf (dorsal view; caudal facing left), n=6. GFP+/RFP+ brain vessels, where GFP marks native endothelial cells and RFP endothelial cells overexpressing human WT-PPIL4. a-e, showing GFP-tagged kdrl expressing embryonic endothelial cells and cerebral vessels. a’-e’, PPIL4 overexpression is restricted to endothelial cells. a’’-e’’, Overlay of red and green channels. a’’’-e’’’, Vascular tracing performed using Imaris Filament application. f, Volcano plot representing differentially expressed genes in RNA-sequencing results of brain enriched endothelial cells upon abrogation of ppil4 (2.5 hpf). Dots represent genes and colors indicate Log2 fold change (FC) and FDR thresholds with genes meeting both (red), those over Log2FC (blue), those meeting FDR (green), or neither (grey). Horizontal and vertical dashed lines indicate, respectively, thresholds for significance (FDR <0.05) and Log2 FC>0.5. See methods for details of the experiment. g,h, Significant terms from the GO-Cellular Component for significantly differentially downregulated (g) and upregulated (h) genes. Dashed lines showing threshold for significance (FDR=0.05). Scale bar: 50 μm.
Extended Data Fig. 8 Genes with strong positive statistical co-dependency with PPIL4 expression are enriched in brain arterial endothelial cells and Wnt signaling pathway.
Analysis of publicly available scRNAseq mouse brain endothelial cell datasets obtained from Vanlandewijck et al. & He, L. et al. (PMID: 29443965). a, PHATE plot identifies two major groups in brain endothelial cells, arterial and venous (top left panel). Pseudotime analysis demonstrating PPIL4 expression predominantly in arterial endothelial cells when compared to venous (SLC38A5) and arterial (GKN3) markers. b, GSEA revealing that the top 50 genes associated with venous endothelial cells are significantly enriched among the top ranked genes specifically expressed in Cluster 0. Similarly, GSEA showing significant enrichment of the top 50 arterial endothelial genes among the top ranked genes in Cluster 1. c, Expression of 200 genes with top knn-DREMI score (y axis) ordered by DREVI-based clustering and by peak expression along PPIL4 (x axis). d, Bar plots showing significantly enriched KEGG pathways for genes above 95th percentile knn-DREMI score and positive relationship with PPIL4.
Extended Data Fig. 9 PPIL4-WT binding to JMJD6 in both nucleus and cytoplasm and overlapping phenotypes in ppil4−/− and jmjd6−/− zebrafish embryos.
a, Lysates of HEK293 cells expressing HA-JMJD6, V5-PPIL4WT and V5-PPIL4G132S were separated into cytoplasmic and nuclear fractions. Cell lysate (input) and V5-IP of cytoplasmic and nuclear fractions were subjected to immunoblotting with anti-HA, anti-V5, lamin A/C (nuclear fraction [right]), tubulin (cytoplasmic fraction [left]) and actin antibodies. The blots shown are representative of three biological replicates. b,c Bright-field images showing necrosis in the head of jmjd6−/− 2.5 dpf embryos. d, Location of sgRNA designed to target jmjd6 leading to a 2bp deletion in exon 3 and a premature stop codon. e-g, Maximum intensity projection (MIP) of representative confocal z-stack images of 2.5 dpf wild type (n=16), jmjd6−/− (n=4), and ppil4−/− (n=15) embryos in the tg(kdrl:gfp)zn1 background (top panels). Brain vessel segmentation in same larvae obtained by Imaris software; colors represent vessel diameter (bottom panels). Homozygous deletion of jmjd6 or ppil4 individually results in dramatic reduction in midbrain CtA complexity and impairment in vascular morphology. Scale bar: 200 μm in b and c; 50 μm in e-g.
Extended Data Fig. 10 ppil4 depletion leads to impaired activation of Wnt signaling in brain parenchyma and brain ECs of 30 hpf zebrafish.
a-c, Maximum intensity projection (MIP) of confocal z-stack images of three representative 30 hpf ppil4−/− and d-f, ppil4+/+ embryos in double transgenic tg(kdrl:gfp; 7xTCF-Xla.Siam:nlsmCherry) background to visualize Wnt signaling activity (red) and endothelial cells (green) (dorsal view and caudal facing up). TCF reporter signal is quantified using the Spots application in Imaris demonstrating loss of TCF reporting cells in brain parenchyma as well as in midbrain CtAs of ppil4−/− embryos. Endothelial specific Wnt-activity is calculated using Spots-Mask for GFP channel in the designated area in a. See methods for details of image processing and presentation. g-i, Quantification of number of TCF reporting cells using the Spots application in Imaris software showing significant decrease in Wnt-activity in ppil4−/− embryos, in (g) overall brain and (h) brain endothelial cells compared with wild type. (i) Wnt signaling activity in overall brain after subtracting the Wnt activity in endothelial cells. n= 4, and 9 embryos for ppil4+/+, and ppil4−/− respectively. Individual values presented with scatter dot plot and median for all quantifications. Statistical tests performed: Two-tailed Student t-test. Scale bar: 100 μm.
Supplementary information
Supplementary Information
Supplementary Figs. 1–6
Supplementary Table 1
Co-segregating variants in all five affected members of the IA200 family. General gnomAD MAF < 0.005
Supplementary Table 2
Coding mutations of PPIL4 in the replication IA cohort
Supplementary Table 3
Clinical Information of IA cases in our cohort carrying rare PPIL4 mutations
Supplementary Table 4
Burden analysis for 17 co-segregating genes in affected individuals of the IA200 family (gnomAD NFE European MAF < 0.0001; LoF + D-Mis (CADD ≥ 30))
Supplementary Table 5
Expression percentile rank of the genes in brain-enriched endothelial cells of 2.5-dpf wild-type zebrafish
Supplementary Table 6
Differential gene expression analysis in RNA-seq between the FACS-sorted brain-enriched endothelial cells of 2.5-dpf-old ppil4−/− and wild-type zebrafish
Supplementary Table 7
Enrichment analysis for the significantly differentially downregulated genes (FDR < 0.05) in RNA-seq between the FACS-sorted brain-enriched endothelial cells of 2.5-dpf-old ppil4−/− and wild-type zebrafish using Metascape (GO-Cellular Compartments)
Supplementary Table 8
Enrichment analysis for the significantly differentially upregulated genes (FDR < 0.05) in RNA-seq between the FACS-sorted brain-enriched endothelial cells of 2.5-dpf-old ppil4−/− and wild-type zebrafish using Metascape (GO-Cellular Compartments)
Supplementary Table 9
knn-DREMI analysis for Ppil4 in publicly available (ref. 27) scRNA-seq data of mouse brain vascular endothelial cells. Cluster 0 and Cluster 2 indicate genes showing positive relationship with Ppil4 expression, whereas Clusters 1, 3 and 4 have negative relationship
Supplementary Table 10
Mean difference analysis in publicly available (ref. 27) scRNA-seq data of mouse brain vascular endothelial cells showing two major clusters: Cluster 0-venous and Cluster 1-arterial
Supplementary Table 11
Enrichment analysis of knn-DREMI top-scored genes (95th percentile) positively regulated with Ppil4 expression in mouse brain endothelial cells using KEGG pathways (Molecular Signatures Database)
Source data
Source Data Fig. 1
Original gel images for Fig. 5
Source Data Fig. 2
Original gel images for Extended Data Fig. 9
Rights and permissions
About this article
Cite this article
Barak, T., Ristori, E., Ercan-Sencicek, A.G. et al. PPIL4 is essential for brain angiogenesis and implicated in intracranial aneurysms in humans. Nat Med 27, 2165–2175 (2021). https://doi.org/10.1038/s41591-021-01572-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-021-01572-7
This article is cited by
-
Serum Interleukin-1 Levels Are Associated with Intracranial Aneurysm Instability
Translational Stroke Research (2024)
-
CCIVR2 facilitates comprehensive identification of both overlapping and non-overlapping antisense transcripts within specified regions
Scientific Reports (2023)
-
Shaping the brain vasculature in development and disease in the single-cell era
Nature Reviews Neuroscience (2023)
-
Critical Roles of Protein Arginine Methylation in the Central Nervous System
Molecular Neurobiology (2023)
-
A Future Blood Test to Detect Cerebral Aneurysms
Cellular and Molecular Neurobiology (2023)