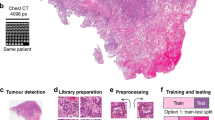
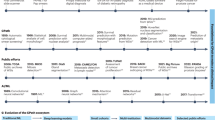
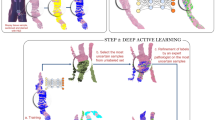
Abstract
Machine learning techniques have great potential to improve medical diagnostics, offering ways to improve accuracy, reproducibility and speed, and to ease workloads for clinicians. In the field of histopathology, deep learning algorithms have been developed that perform similarly to trained pathologists for tasks such as tumor detection and grading. However, despite these promising results, very few algorithms have reached clinical implementation, challenging the balance between hope and hype for these new techniques. This Review provides an overview of the current state of the field, as well as describing the challenges that still need to be addressed before artificial intelligence in histopathology can achieve clinical value.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Yu, K. H., Beam, A. L. & Kohane, I. S. Artificial intelligence in healthcare. Nat. Biomed. Eng. 2, 719–731 (2018).
Hamet, P. & Tremblay, J. Artificial intelligence in medicine. Metabolism 69, S36–S40 (2017).
Litjens, G. et al. A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017).
Bulten, W. et al. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: a diagnostic study. Lancet Oncol. 21, 233–241 (2020).
Ehteshami Bejnordi, B. et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318, 2199–2210 (2017).
Bera, K., Schalper, K. A., Rimm, D. L., Velcheti, V. & Madabhushi, A. Artificial intelligence in digital pathology – new tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 16, 703–715 (2019).
Fuchs, T. J. & Buhmann, J. M. Computational pathology: challenges and promises for tissue analysis. Comput. Med. Imaging Graph. 35, 515–530 (2011).
Louis, D. N. et al. Computational pathology: an emerging definition. Arch. Pathol. Lab. Med. 138, 1133–1138 (2014).
Mendelsohn, M. L., Kolman, W. A., Perry, B. & Prewitt, J. M. Computer analysis of cell images. Postgrad. Med. 38, 567–573 (1965).
Zwanenburg, A. et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 295, 328–338 (2020).
Beck, A. H. et al. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci. Transl. Med. 3, 108–113 (2011).
Kather, J. N. et al. Multi-class texture analysis in colorectal cancer histology. Sci. Rep. 6, 27988 (2016).
Madabhushi, A. & Lee, G. Image analysis and machine learning in digital pathology: challenges and opportunities. Med. Image Anal. 33, 170–175 (2016).
Srinidhi, C. L., Ciga, O. & Martel, A. L. Deep neural network models for computational histopathology: a survey. Med. Image Anal. 67, 101813 (2021).
Cireşan, D. C., Meier, U., Masci, J., Gambardella, L. M. & Schmidhuber, J. Flexible, high performance convolutional neural networks for image classification. In Proc. 22nd International Joint Conference on Artificial Intelligence 1237–1242 (2011).
Cireşan, D. C., Giusti, A., Gambardella, L. M. & Schmidhuber, J. Mitosis detection in breast cancer histology images with deep neural networks. In Proc. Medical Image Computing and Computer-Assisted Intervention, Lecture Notes in Computer Science Vol. 8150, 411–418 (2013).
Cruz-Roa, A. et al. Automatic detection of invasive ductal carcinoma in whole slide images with convolutional neural networks. In Proc. SPIE Medical Imaging Vol. 9041, 904103 (2014).
Ertosun, M. G. & Rubin, D. L. Automated grading of gliomas using deep learning in digital pathology images: a modular approach with ensemble of convolutional neural networks. In Proc. American Medical Informatics Association Annual Symposium 1899–1908 (2015).
Wong, G. L. et al. Artificial intelligence in prediction of non-alcoholic fatty liver disease and fibrosis. J. Gastroenterol. Hepatol. 36, 543–550 (2021).
Hermsen, M. et al. Deep learning–based histopathologic assessment of kidney tissue. J. Am. Soc. Nephrol. 30, 1968–1979 (2019).
Litjens, G. et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci. Rep. 6, 26286 (2016).
Litjens, G. et al. 1399 H&E-stained sentinel lymph node sections of breast cancer patients: the CAMELYON dataset. GigaScience 7, giy065 (2018).
Liu, Y. et al. Detecting cancer metastases on gigapixel pathology images. Preprint at https://arxiv.org/abs/1703.02442 (2017).
White House Office of Science and Technology Policy. Preparing for the Future of Artificial Intelligence (2016); https://obamawhitehouse.archives.gov/sites/default/files/whitehouse_files/microsites/ostp/NSTC/preparing_for_the_future_of_ai.pdf
Wang, D., Khosla, A., Gargeya, R., Irshad, H. & Beck, A. H. Deep learning for identifying metastatic breast cancer. Preprint at https://arxiv.org/abs/1606.05718 (2016).
Wang, X. et al. Weakly supervised deep learning for whole slide lung cancer image analysis. IEEE Trans. Cybern. 50, 3950–3962 (2019).
Syrykh, C. et al. Accurate diagnosis of lymphoma on whole-slide histopathology images using deep learning. NPJ Digital Med. 3, 63 (2020).
Tabibu, S., Vinod, P. K. & Jawahar, C. V. Pan-renal cell carcinoma classification and survival prediction from histopathology images using deep learning. Sci. Rep. 9, 10509 (2019).
Sari, C. T. & Gunduz-Demir, C. Unsupervised feature extraction via deep learning for histopathological classification of colon tissue images. IEEE Trans. Med. Imaging 38, 1139–1149 (2019).
Rawat, R. R. et al. Deep learned tissue ‘fingerprints’ classify breast cancers by ER/PR/Her2 status from H&E images. Sci. Rep. 10, 7275 (2020).
Lee, B. & Paeng, K. A robust and effective approach towards accurate metastasis detection and pN-stage classification in breast cancer. In Proc. Medical Image Computing and Computer Assisted Intervention, Lecture Notes in Computer Science Vol. 11071, 841–850 (2018).
Awan, R., Koohbanani, N. A., Shaban, M., Lisowska, A. & Rajpoot, N. Context-aware learning using transferable features for classification of breast cancer histology images. In Proc. International Conference on Image Analysis and Recognition 788–795 (2018).
Feng, Y., Zhang, L. & Mo, J. Deep manifold preserving autoencoder for classifying breast cancer histopathological images. IEEE/ACM Trans. Comput. Biol. Bioinform. 17, 91–101 (2020).
Galateau Salle, F. et al. Comprehensive molecular and pathologic evaluation of transitional mesothelioma assisted by deep learning approach: a multi-institutional study of the international mesothelioma panel from the MESOPATH Reference Center. J. Thorac. Oncol. 15, 1037–1053 (2020).
Iizuka, O. et al. Deep learning models for histopathological classification of gastric and colonic epithelial tumours. Sci. Rep. 10, 1504 (2020).
Kiani, A. et al. Impact of a deep learning assistant on the histopathologic classification of liver cancer. NPJ Digital Med. 3, 23 (2020).
Kwok, S. Multiclass classification of breast cancer in whole-slide images. In Proc. International Conference on Image Analysis and Recognition 931–940 (2018).
Wei, J. W. et al. Pathologist-level classification of histologic patterns on resected lung adenocarcinoma slides with deep neural networks. Sci. Rep. 9, 3358 (2019).
Yang, H., Kim, J. Y., Kim, H. & Adhikari, S. P. Guided soft attention network for classification of breast cancer histopathology images. IEEE Trans. Med. Imaging 39, 1306–1315 (2020).
Pinckaers, H. & Litjens, G. Neural ordinary differential equations for semantic segmentation of individual colon glands. Preprint at https://arxiv.org/abs/1910.10470 (2019).
Naylor, P., Lae, M., Reyal, F. & Walter, T. Segmentation of nuclei in histopathology images by deep regression of the distance map. IEEE Trans. Med. Imaging 38, 448–459 (2019).
Long, F. Microscopy cell nuclei segmentation with enhanced U-Net. BMC Bioinformatics 21, 8 (2020).
Jia, Z., Huang, X., Chang, E. I.-C. & Xu, Y. Constrained deep weak supervision for histopathology image segmentation. IEEE Trans. Med. Imaging 36, 2376–2388 (2017).
Graham, S. et al. MILD-Net: minimal information loss dilated network for gland instance segmentation in colon histology images. Med. Image Anal. 52, 199–211 (2019).
Graham, S. et al. Hover-Net: simultaneous segmentation and classification of nuclei in multi-tissue histology images. Med. Image Anal. 58, 101563 (2019).
Agarwalla, A., Shaban, M. & Rajpoot, N. M. Representation-aggregation networks for segmentation of multi-gigapixel histology images. Preprint at https://arxiv.org/abs/1707.08814 (2017).
Bueno, G., Fernandez-Carrobles, M. M., Gonzalez-Lopez, L. & Deniz, O. Glomerulosclerosis identification in whole slide images using semantic segmentation. Comput. Methods Prog. Biomed. 184, 105273 (2020).
Chen, H. et al. DCAN: deep contour-aware networks for object instance segmentation from histology images. Med. Image Anal. 36, 135–146 (2017).
de Bel, T. et al. Automatic segmentation of histopathological slides of renal tissue using deep learning. in Proc. SPIE Medical Imaging Digital Pathology, 1058112 (2018); https://doi.org/10.1117/12.2293717
Xu, G. et al. CAMEL: a weakly supervised learning framework for histopathology image segmentation. In Proc. International Conference on Computer Vision 10681–10690 (2019).
Sirinukunwattana, K. et al. Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans. Med. Imaging 35, 1196–1206 (2016).
Swiderska-Chadaj, Z. et al. Learning to detect lymphocytes in immunohistochemistry with deep learning. Med. Image Anal. 58, 101547 (2019).
Le, H. et al. Utilizing automated breast cancer detection to identify spatial distributions of tumor-infiltrating lymphocytes in invasive breast cancer. Am. J. Pathol. 190, 1491–1504 (2020).
Akbar, S. et al. Automated and manual quantification of tumour cellularity in digital slides for tumour burden assessment. Sci. Rep. 9, 14099 (2019).
Hou, L. et al. Sparse autoencoder for unsupervised nucleus detection and representation in histopathology images. Pattern Recognit. 86, 188–200 (2019).
Veta, M. et al. Predicting breast tumor proliferation from whole-slide images: the TUPAC16 challenge. Med. Image Anal. 54, 111–121 (2019).
Tellez, D. et al. Whole-slide mitosis detection in H&E breast histology using PHH3 as a reference to train distilled stain-invariant convolutional networks. IEEE Trans. Med. Imaging 37, 2126–2136 (2018).
Mahmood, T., Arsalan, M., Owais, M., Lee, M. B. & Park, K. R. Artificial intelligence-based mitosis detection in breast cancer histopathology images using faster R-CNN and deep CNNs. J. Clin. Med. 9, 749 (2020).
Chen, H., Wang, X. & Heng, P. A. Automated mitosis detection with deep regression networks. In Proc. IEEE International Symposium on Biomedical Imaging 1204–1207 (2016).
Li, C. et al. Weakly supervised mitosis detection in breast histopathology images using concentric loss. Med. Image Anal. 53, 165–178 (2019).
Nagpal, K. et al. Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. NPJ Digital Med. 2, 48 (2019).
Jansen, I. et al. Automated detection and grading of non-muscle-invasive urothelial cell carcinoma of the bladder. Am. J. Pathol. 190, 1483–1490 (2020).
Karimi, D. et al. Deep learning-based Gleason grading of prostate cancer from histopathology images—role of multiscale decision aggregation and data augmentation. IEEE J. Biomed. Health Inform. 24, 1413–1426 (2020).
Korde, L. A. et al. Neoadjuvant chemotherapy, endocrine therapy, and targeted therapy for breast cancer: ASCO guideline. J. Clin. Oncol. https://doi.org/10.1200/JCO.20.03399 (2021).
Balkenhol, M. et al. Deep learning assisted mitotic counting for breast cancer. Lab. Invest. 99, 1596–1606 (2019).
Ström, P. et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: a population-based, diagnostic study. Lancet Oncol. 21, 222–232 (2020).
Gertych, A. et al. Convolutional neural networks can accurately distinguish four histologic growth patterns of lung adenocarcinoma in digital slides. Sci. Rep. 9, 1483 (2019).
Salgado, R. et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann. Oncol. 26, 259–271 (2015).
Saltz, J. et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 23, 181–193 (2018).
AbdulJabbar, K. Geospatial immune variability illuminates differential evolution of lung adenocarcinoma. Nat. Med. 26, 1054–1062 (2020).
Kather, J. N. et al. Predicting survival from colorectal cancer histology slides using deep learning: a retrospective multicenter study. PLoS Med. 16, e1002730 (2019).
Geessink, O. G. F. et al. Computer aided quantification of intratumoral stroma yields an independent prognosticator in rectal cancer. Cell. Oncol. 42, 331–341 (2019).
Kapil, A. et al. DASGAN–joint domain adaptation and segmentation for the analysis of epithelial regions in histopathology PD-L1 images. Preprint at https://arxiv.org/abs/1906.11118 (2019).
Sha, L. et al. Multi-field-of-view deep learning model predicts non small cell lung cancer programmed death-ligand 1 status from whole-slide hematoxylin and eosin images. J. Pathol. Inform. 10, 24 (2019).
Campanella, G. et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 25, 1301–1309 (2019).
Zhang, Z. et al. Pathologist-level interpretable whole-slide cancer diagnosis with deep learning. Nat. Mach. Intell. 1, 236–245 (2019).
Zhou, C. et al. Histopathology classification and localization of colorectal cancer using global labels by weakly supervised deep learning. Comput. Med. Imaging Graph. 88, 101861 (2021).
Song, Z. et al. Clinically applicable histopathological diagnosis system for gastric cancer detection using deep learning. Nat. Commun. 11, 4294 (2020).
Albarqouni, S. et al. AggNet: deep learning from crowds for mitosis detection in breast cancer histology images. IEEE Trans. Med. Imaging 35, 1313–1321 (2016).
Amgad, M. et al. Structured crowdsourcing enables convolutional segmentation of histology images. Bioinformatics 35, 3461–3467 (2019).
Bulten, W. et al. Epithelium segmentation using deep learning in H&E-stained prostate specimens with immunohistochemistry as reference standard. Sci. Rep. 9, 864 (2019).
Valkonen, M. et al. Cytokeratin-supervised deep learning for automatic recognition of epithelial cells in breast cancers stained for ER, PR, and Ki-67. IEEE Trans. Med. Imaging 39, 534–542 (2020).
Alemi Koohbanani, N., Jahanifar, M., Zamani Tajadin, N. & Rajpoot, N. NuClick: a deep learning framework for interactive segmentation of microscopic images. Med. Image Anal. 65, 101771 (2020).
Bokhorst, J. M. et al. Learning from sparsely annotated data for semantic segmentation in histopathology images. In Proc. International Conference on Medical Imaging with Deep Learning, Proceedings of Machine Learning Research Vol. 102, 84–91 (2019).
Brieu, N. et al. Domain adaptation-based augmentation for weakly supervised nuclei detection. Preprint at https://arxiv.org/abs/1907.04681 (2019).
Gadermayr, M., Gupta, L., Klinkhammer, B. M., Boor, P. & Merhof, D. Unsupervisedly training GANs for segmenting digital pathology with automatically generated annotations. In Proc. International Conference on Medical Imaging with Deep Learning, Proceedings of Machine Learning Research Vol. 102, 175–184 (2019).
Liang, Q. et al. Weakly supervised biomedical image segmentation by reiterative learning. IEEE J. Biomed. Health Inform. 23, 1205–1214 (2019).
Gao, S. et al. Using case-level context to classify cancer pathology reports. PLoS ONE 15, e0232840 (2020).
Alawad, M. et al. Automatic extraction of cancer registry reportable information from free-text pathology reports using multitask convolutional neural networks. J. Am. Med. Inform. Assoc. 27, 89–98 (2020).
Coudray, N. et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 24, 1559–1567 (2018).
Pawlowski, N. et al. Needles in haystacks: on classifying tiny objects in large images. Preprint at https://arxiv.org/abs/1908.06037 (2019).
Ilse, M., Tomczak, J. M. & Welling, M. Attention-based deep multiple instance learning. In Proc. International Conference on Machine Learning, Proceedings of Machine Learning Research Vol. 80, 2127–2136 (2018).
Hou, L. et al. Patch-based convolutional neural network for whole slide tissue image classification. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 2424–2433 (2016).
Lu, M. Y. et al. Data efficient and weakly supervised computational pathology on whole slide images. Nat. Biomed. Eng. https://doi.org/10.1038/s41551-020-00682-w (2021).
Tellez, D., Litjens, G., van der Laak, J. & Ciompi, F. Neural image compression for gigapixel histopathology image analysis. IEEE Trans. Pattern Anal. Mach. Intell. 43, 567–578 (2021).
Pinckaers, J. H. F. M., van Ginneken, B. & Litjens, G. Streaming convolutional neural networks for end-to-end learning with multi-megapixel images. IEEE Trans. Pattern Anal. Mach. Intell. https://doi.org/10.1109/TPAMI.2020.3019563 (2020).
Wulczyn, E. et al. Deep learning-based survival prediction for multiple cancer types using histopathology images. PLoS ONE 15, e0233678 (2020).
Skrede, O. J. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet 395, 350–360 (2020).
Saillard, C. et al. Predicting survival after hepatocellular carcinoma resection using deep-learning on histological slides. Hepatology 72, 2000–2013 (2020).
Qaiser, T. et al. Digital tumor-collagen proximity signature predicts survival in diffuse large B-cell lymphoma. In Proc. European Congress on Digital Pathology, Lecture Notes in Computer Science Vol. 11435, 163–171 (2019).
Mobadersany, P. et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl Acad. Sci. USA 115, E2970–E2979 (2018).
Bychkov, D. et al. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci. Rep. 8, 3395 (2018).
Courtiol, P. et al. Deep learning-based classification of mesothelioma improves prediction of patient outcome. Nat. Med. 25, 1519–1525 (2019).
Kulkarni, P. M. et al. Deep learning based on standard H&E images of primary melanoma tumors identifies patients at risk for visceral recurrence and death. Clin. Cancer Res. 26, 1126–1134 (2020).
Cui, D., Liu, Y., Liu, G. & Liu, L. A multiple-instance learning-based convolutional neural network model to detect the IDH1 mutation in the histopathology images of glioma tissues. J. Comput. Biol. 27, 1264–1272 (2020).
Liu, S. et al. Isocitrate dehydrogenase (IDH) status prediction in histopathology images of gliomas using deep learning. Sci. Rep. 10, 7733 (2020).
Kather, J. N. et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 25, 1054–1056 (2019).
Kather, J. N. et al. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat. Cancer 1, 789–799 (2020).
Fu, Y. et al. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nat. Cancer 1, 800–810 (2020).
Schmauch, B. et al. A deep learning model to predict RNA-Seq expression of tumours from whole slide images. Nat. Commun. 11, 3877 (2020).
Couture, H. D. et al. Image analysis with deep learning to predict breast cancer grade, ER status, histologic subtype, and intrinsic subtype. NPJ Breast Cancer 4, 30 (2018).
Sirinukunwattana, K. et al. Image-based consensus molecular subtype (imCMS) classification of colorectal cancer using deep learning. Gut 70, 544–554 (2021).
Durán, J. M. & Jongsma, K. R. Who is afraid of black box algorithms? On the epistemological and ethical basis of trust in medical AI. J. Med Ethics https://doi.org/10.1136/medethics-2020-106820 (2021).
Abels, E. et al. Computational pathology definitions, best practices, and recommendations for regulatory guidance: a white paper from the Digital Pathology Association. J. Pathol. 249, 286–294 (2019).
Kelly, C. J., Karthikesalingam, A., Suleyman, M., Corrado, G. & King, D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 17, 195 (2019).
McKinney, S. M. et al. International evaluation of an AI system for breast cancer screening. Nature 577, 89–94 (2020).
Panch, T., Mattie, H. & Atun, R. Artificial intelligence and algorithmic bias: implications for health systems. J. Glob. Health 9, 010318 (2019).
de Bel, T., Hermsen, M., Kers, J., van der Laak, J. & Litjens, G. J. S. Stain-transforming cycle-consistent generative adversarial networks for improved segmentation of renal histopathology. In Proc. International Conference on Medical Imaging with Deep Learning, Proceedings of Machine Learning Research Vol. 102, 151–163 (2019).
Liu, Y. et al. Artificial intelligence-based breast cancer nodal metastasis detection: insights into the black box for pathologists. Arch. Pathol. Lab. Med. 143, 859–868 (2019).
Tellez, D. et al. Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology. Med. Image Anal. 58, 101544 (2019).
Cho, H., Lim, S., Choi, G. & Min, H. Neural stain-style transfer learning using GAN for histopathological images. Preprint at https://arxiv.org/abs/1710.08543 (2017).
Janowczyk, A., Basavanhally, A. & Madabhushi, A. Stain Normalization using Sparse AutoEncoders (StaNoSA): application to digital pathology. Comput. Med. Imaging Graph. 57, 50–61 (2017).
Shaban, M. T., Baur, C., Navab, N. & Albarqouni, S. StainGAN: stain style transfer for digital histological images. In Proc. IEEE International Symposium on Biomedical Imaging 953–956 (2019).
Zheng, Y. et al. Stain standardization capsule for application-driven histopathological image normalization. IEEE J. Biomed. Health Inform. 25, 337–347 (2021).
Linmans, J., van der Laak, J. & Litjens, G. Efficient out-of-distribution detection in digital pathology using multi-head convolutional neural networks. In Proc. Conference on Medical Imaging with Deep Learning, Proceedings of Machine Learning Research Vol. 121, 465–478 (2020).
Kohl, S. et al. A probabilistic U-Net for segmentation of ambiguous images. Adv. Neural Inf. Process. Syst. (2018).
Kleppe, A. et al. Designing deep learning studies in cancer diagnostics. Nat. Rev. Cancer 21, 199–211 (2021).
Staartjes, V. E. & Kernbach, J. M. Significance of external validation in clinical machine learning: let loose too early. Spine J. 20, 1159–1160 (2020).
Beede, E. et al. A human-centered evaluation of a deep learning system deployed in clinics for the detection of diabetic retinopathy. In Proc. CHI Conference on Human Factors in Computing Systems 1–12 (2020).
Dudgeon, S. N. et al. A pathologist-annotated dataset for validating artificial intelligence: a project description and pilot study. Preprint at https://arxiv.org/abs/2010.06995 (2020).
Nagendran, M. et al. Artificial intelligence versus clinicians: systematic review of design, reporting standards, and claims of deep learning studies. BMJ 368, m689 (2020).
Allen, T. C. Regulating artificial intelligence for a successful pathology future. Arch. Pathol. Lab. Med. 143, 1175–1179 (2019).
Dong, J. et al. Clinical trials for artificial intelligence in cancer diagnosis: a cross-sectional study of registered trials in ClinicalTrials.gov. Front. Oncol. 15, 1629 (2020).
Collins, G. S. & Moons, K. G. M. Reporting of artificial intelligence prediction models. Lancet 393, 1577–1579 (2019).
Chen, P. H. C. et al. An augmented reality microscope with real-time artificial intelligence integration for cancer diagnosis. Nat. Med. 25, 1453–1457 (2019).
Guidotti, R. et al. A survey of methods for explaining black box models. ACM Comput. Surv. 51, 93 (2019).
Kroll, J. A. The fallacy of inscrutability. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 376, 20180084 (2018).
US Food and Drug Administration (FDA). Proposed Regulatory Framework for Modifications to Artificial Intelligence/Machine Learning (AI/ML)-Based Software as a Medical Device (SaMD)—Discussion Paper and Request for Feedback. https://www.fda.gov/files/medicaldevices/published/US-FDA-Artificial-Intelligence-and-Machine-Learning-Discussion-Paper.pdf (accessed 3 May, 2021).
Price, W. N. & Cohen, I. G. Privacy in the age of medical big data. Nat. Med. 25, 37–43 (2019).
Laï, M. C., Brian, M. & Mamzer, M. F. Perceptions of artificial intelligence in healthcare: findings from a qualitative survey study among actors in France. J. Transl. Med. 18, 14 (2020).
Rieke, N. et al. The future of digital health with federated learning. NPJ Digit. Med. 3, 119 (2020).
Sheller, M. J. et al. Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data. Sci. Rep. 10, 12598 (2020).
European Commission. Ethics Guidelines for Trustworthy AI (2019); https://ec.europa.eu/digital-single-market/en/news/ethics-guidelines-trustworthy-ai
Sirinukunwattana, K. et al. Gland segmentation in colon histology images: the glas challenge contest. Med. Image Anal. 35, 489–502 (2017).
Ronneberger, O., Fischer, P. & Brox, T. U-Net: convolutional networks for biomedical image segmentation. In Proc. Medical Image Computing and Computer-Assisted Intervention, Lecture Notes in Computer Science Vol. 9351, 234–241 (2015).
Acknowledgements
We thank M. Hermsen for providing Fig. 1. J.v.d.L. acknowledges funding from the Knut and Alice Wallenberg Foundation, Sweden, and received funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement no. 945358. This Joint Undertaking receives support from the European Union’s Horizon 2020 research and innovation program and EFPIA. G.L. acknowledges funding from the Dutch Cancer Society (KUN 2015-7970) and the Netherlands Organization for Scientific Research (NWO; project number 016.186.152). F.C. acknowledges funding from the European Union’s Horizon 2020 research and innovation program under grant agreement no. 825292 (ExaMode, http://www.examode.eu/); the Dutch Cancer Society (KWF; project no. 11917); and the Netherlands Organization for Scientific Research (NWO; project no. 18388).
Author information
Authors and Affiliations
Contributions
All authors were involved in identifying relevant literature, and in drafting and revising the manuscript.
Corresponding author
Ethics declarations
Competing interests
J.v.d.L. is a member of the advisory boards of Philips, the Netherlands, and ContextVision, Sweden, and received research funding from Philips, the Netherlands, ContextVision, Sweden, and Sectra, Sweden, in the last 5 years. G.L. is a member of the Medical Advances Advisory Board of Vital Images (Minnetonka, USA), received research funding from Philips Digital Pathology Solutions (Best, the Netherlands) and had a consultancy role for Novartis (Basel, Switzerland). F.C. is chair of the Scientific and Medical Advisory Board of TRIBVN Healthcare (Paris, France).
Additional information
Peer review information Nature Medicine thanks Richard Levenson, Nasir Rajpoot and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Joao Monteiro was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
van der Laak, J., Litjens, G. & Ciompi, F. Deep learning in histopathology: the path to the clinic. Nat Med 27, 775–784 (2021). https://doi.org/10.1038/s41591-021-01343-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-021-01343-4
This article is cited by
-
Slideflow: deep learning for digital histopathology with real-time whole-slide visualization
BMC Bioinformatics (2024)
-
Digital image analysis and machine learning-assisted prediction of neoadjuvant chemotherapy response in triple-negative breast cancer
Breast Cancer Research (2024)
-
Algorithm-assisted diagnosis of Hirschsprung’s disease – evaluation of robustness and comparative image analysis on data from various labs and slide scanners
Diagnostic Pathology (2024)
-
tRigon: an R package and Shiny App for integrative (path-)omics data analysis
BMC Bioinformatics (2024)
-
Open and reusable deep learning for pathology with WSInfer and QuPath
npj Precision Oncology (2024)