Abstract
Langerhans cell histiocytosis (LCH) is a potentially fatal condition characterized by granulomatous lesions with characteristic clonal mononuclear phagocytes (MNPs) harboring activating somatic mutations in mitogen-activated protein kinase (MAPK) pathway genes, most notably BRAFV600E. We recently discovered that the BRAFV600E mutation can also affect multipotent hematopoietic progenitor cells (HPCs) in multisystem LCH disease. How the BRAFV600E mutation in HPCs leads to LCH is not known. Here we show that enforced expression of the BRAFV600E mutation in early mouse and human multipotent HPCs induced a senescence program that led to HPC growth arrest, apoptosis resistance and a senescence-associated secretory phenotype (SASP). SASP, in turn, promoted HPC skewing toward the MNP lineage, leading to the accumulation of senescent MNPs in tissue and the formation of LCH lesions. Accordingly, elimination of senescent cells using INK-ATTAC transgenic mice, as well as pharmacologic blockade of SASP, improved LCH disease in mice. These results identify senescent cells as a new target for the treatment of LCH.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Datasets supporting the findings presented in this study are available from the corresponding author upon reasonable request. Any data that can be shared will be released via a material transfer agreement. Microarray sequencing data obtained from CD34+ BM cells transduced with BRAFV600E or control NGFR lentiviral vector are listed in Supplementary Table 3. RNA-seq data obtained from CD34+ cells purified from the BM of patients with LCH and healthy donors are listed in Supplementary Table 4. Source data are provided with this paper.
References
Merad, M., Ginhoux, F. & Collin, M. Origin, homeostasis and function of Langerhans cells and other langerin-expressing dendritic cells. Nat. Rev. Immunol. 8, 935–947 (2008).
Stålemark, H. et al. Incidence of Langerhans cell histiocytosis in children: a population-based study. Pediatr. Blood Cancer 51, 76–81 (2008).
Allen, C. E., Merad, M. & McClain, K. L. Langerhans-cell histiocytosis. N. Engl. J. Med. 379, 856–868 (2018).
Badalian-Very, G. et al. Recurrent BRAF mutations in Langerhans cell histiocytosis. Blood 116, 1919–1923 (2010).
Chakraborty, R. et al. Mutually exclusive recurrent somatic mutations in MAP2K1 and BRAF support a central role for ERK activation in LCH pathogenesis. Blood 124, 3007–3015 (2014).
Chakraborty, R. et al. Alternative genetic mechanisms of BRAF activation in Langerhans cell histiocytosis. Blood 128, 2533–2537 (2016).
Lim, K. P. H. et al. Circulating CD1c+ myeloid dendritic cells are potential precursors to LCH lesion CD1a+CD207+ cells. Blood Adv. 4, 87–99 (2020).
Berres, M.-L. et al. BRAF-V600E expression in precursor versus differentiated dendritic cells defines clinically distinct LCH risk groups. J. Exp. Med. 211, 669–683 (2014).
Durham, B. H. et al. Functional evidence for derivation of systemic histiocytic neoplasms from hematopoietic stem/progenitor cells. Blood 130, 176–180 (2017).
Milne, P. et al. Hematopoietic origin of Langerhans cell histiocytosis and Erdheim–Chester disease in adults. Blood 130, 167–175 (2017).
Berres, M.-L., Merad, M. & Allen, C. E. Progress in understanding the pathogenesis of Langerhans cell histiocytosis: back to Histiocytosis X? Br. J. Haematol. 169, 3–13 (2015).
Göthert, J. R. et al. In vivo fate-tracing studies using the Scl stem cell enhancer: embryonic hematopoietic stem cells significantly contribute to adult hematopoiesis. Blood 105, 2724–2732 (2005).
Michaloglou, C. et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436, 720–724 (2005).
Hernandez-Segura, A., Nehme, J. & Demaria, M. Hallmarks of cellular senescence. Trends Cell Biol. 28, 436–453 (2018).
Muñoz-Espín, D. & Serrano, M. Cellular senescence: from physiology to pathology. Nat. Rev. Mol. Cell Biol. 15, 482–496 (2014).
Kuilman, T. et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 133, 1019–1031 (2008).
McClain, K. L. et al. CNS Langerhans cell histiocytosis: common hematopoietic origin for LCH-associated neurodegeneration and mass lesions. Cancer 124, 2607–2620 (2018).
Herranz, N. et al. mTOR regulates MAPKAPK2 translation to control the senescence-associated secretory phenotype. Nat. Cell Biol. 17, 1205–1217 (2015).
Laberge, R.-M. et al. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat. Cell Biol. 17, 1049–1061 (2015).
Baker, D. J. et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479, 232–236 (2011).
Wilson, W. H. et al. Navitoclax, a targeted high-affinity inhibitor of BCL-2, in lymphoid malignancies: a phase 1 dose-escalation study of safety, pharmacokinetics, pharmacodynamics, and antitumour activity. Lancet. Oncol. 11, 1149–1159 (2010).
Bussian, T. J. et al. Clearance of senescent glial cells prevents tau-dependent pathology and cognitive decline. Nature 562, 578–582 (2018).
Chang, J. et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 22, 78–83 (2016).
Jeon, O. H. et al. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat. Med. 23, 775–781 (2017).
Hogstad, B. et al. RAF/MEK/extracellular signal-related kinase pathway suppresses dendritic cell migration and traps dendritic cells in Langerhans cell histiocytosis lesions. J. Exp. Med. 215, 319–336 (2018).
Xerri, L. et al. CDKN2A/B deletion and double-hit mutations of the MAPK pathway underlie the aggressive behavior of Langerhans cell tumors. Am. J. Surg. Pathol. 42, 150–159 (2018).
Picarsic, J. & Jaffe, R. Nosology and pathology of Langerhans cell histiocytosis. Hematol. Oncol. Clin. North Am. 29, 799–823 (2015).
McClain, K., Jin, H., Gresik, V. & Favara, B. Langerhans cell histiocytosis: lack of a viral etiology. Am. J. Hematol. 47, 16–20 (1994).
Jenson, H. B., McClain, K. L., Leach, C. T., Deng, J. H. & Gao, S. J. Evaluation of human herpesvirus type 8 infection in childhood Langerhans cell histiocytosis. Am. J. Hematol. 64, 237–241 (2000).
Montecino-Rodriguez, E., Berent-Maoz, B. & Dorshkind, K. Causes, consequences, and reversal of immune system aging. J. Clin. Invest. 123, 958–965 (2013).
Cohen Aubart, F. et al. Targeted therapies in 54 patients with Erdheim–Chester disease, including follow-up after interruption (the LOVE study). Blood 130, 1377–1380 (2017).
Diamond, E. L. et al. Efficacy of MEK inhibition in patients with histiocytic neoplasms. Nature 567, 521–524 (2019).
Eckstein, O. S., Visser, J., Rodriguez-Galindo, C. & Allen, C. E. Clinical responses and persistent BRAF V600E+ blood cells in children with LCH treated with MAPK pathway inhibition. Blood 133, 1691–1694 (2019).
Lee, J. et al. Lineage specification of human dendritic cells is marked by IRF8 expression in hematopoietic stem cells and multipotent progenitors. Nat. Immunol. 18, 877–888 (2017).
Dankort, D. et al. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev. 21, 379–384 (2007).
Baccarini, A. et al. Kinetic analysis reveals the fate of a microRNA following target regulation in mammalian cells. Curr. Biol. 21, 369–376 (2011).
Remark, R. et al. In-depth tissue profiling using multiplexed immunohistochemical consecutive staining on single slide. Sci. Immunol. 1, aaf6925 (2016).
Acknowledgements
We thank the Biorepository and Pathology Core and the Flow Cytometry Core Facilities at the Icahn School of Medicine at Mount Sinai for their technical expertise. We also thank K. Phaik Har Lim, T.-K. Man, T. Burke and B. Scull for their help with the generation of microarray sequencing data. We thank J. Kofler for kindly providing LCH-ND pictures and performing immunostaining. We thank L. Troncoso for her help. We thank C. Woods, image application specialist, Cincinnati Children’s Hospital Medical Center, for figure preparation for human studies (Fig. 4g) and J. Kofler, UPMC Division of Neuropathology, for creating Extended Data Fig. 4e. M.M. received funding from the National Institute of Health (R01 CA154947 and R01 CA190400). C.B. received fellowships from the Fondation pour la Recherche Médicale (FDM20170638478), from l’Institut Servier and from Assistance Publique Hôpitaux de Paris (Année Recherche). C.M.W. is supported by the Swiss National Science Foundation (SNSF PostDoc Mobility Fellowship P400PM_186740) and by the Swiss Cancer League (grant for bursaries BIL KFS 4724-02-2019). We thank the research coordinators of the Histio–Lymphoma team at Texas Children’s Cancer Center for their help with patient sample collection. The TXCH Histiocytosis Program is supported by a research grant from the HistioCure Foundation. This work was supported by the Department of Defense through the Peer Reviewed Cancer Research Program under award no. W81XWH-19-1-0167 (R.C.).
Author information
Authors and Affiliations
Contributions
C.B., J.L.B., C.M.W., R.C., S.T.C., A.T., R.M., H.A., J.J., I.L., M.C.-A., J.C.M., J.G., J.P. and H.A. designed and carried out experiments. A.B., P.I.P., A.L., S.G., K.L.M., J.P., C.E.A. and M.M. supervised experiments and analyzed and interpreted data. C.B., C.E.A. and M.M. drafted the manuscript. C.E.A. and M.M. designed the study, supervised experimental data collection and coordinated integration of collaboration between all participating laboratories. All authors critically reviewed and edited the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Medicine thanks the anonymous reviewers for their contribution to the peer review of this work. Joao Monteiro was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
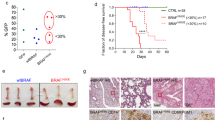
Extended Data Fig. 1 Langerhans Cell Histiocytosis mouse models.
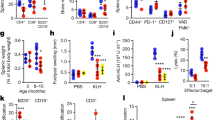
a-f, BRAFV600EScl+ mice and BRAFwtScl+ mice were generated as described in Fig. 1a. a, Representative images of spleen, lung, and femurs at 4 weeks post tamoxifen injections. b, Liver and spleen weights. c, Absolute number of BM cells in BRAFV600EScl+ mice and control littermates (n = 10 mice per group). d, Hematoxylin and eosin staining and CD207 immunohistochemistry staining of tissues isolated from BRAFwtScl+ mice (n = 2–3 mice per group). e, Absolute numbers of lung, dermal and epidermal immune cells in BRAFV600EScl+ mice and control littermates. Data are representative of 3 experiments (n = 3–8). f, Percentage of immune cell populations among lung and skin infiltrating RosaYFP+ cells in BRAFV600EScl+ mice and control littermates. Data are representative of 3 experiments (n = 3–8). g, Scheme of the lentiviral vector constructs used to transduce human CD34+ HPC. h, Graph shows that the transduction efficiency of BRAFV600E and NGFR lentiviral constructs in human CD34+ HPC does not exceed 40 %. i, Western blot of BRAF and phospho-ERK performed on purified BRAFV600Ehu and NGFRhu human CD34+ HPC 7 days after transduction. j, Representative flow plot showing the percentage of CD11c+ CD14+ MNP among circulating blood cells in NSG mice reconstituted with BRAFV600Ehu and NGFRhu HPC. Graph represents the percentage of CD11c and or CD14+ cells among human CD45+ cells from NSG circulating blood (n = 2–3 mice per group). Data are represented as mean ± s.e.m; statistical significance analyzed by an unpaired two-sided t-test is indicated by *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Extended Data Fig. 2 Myeloid skewing of BRAFV600E+ hematopoietic progenitors.
a, Flow cytometry gating strategy for hematopoietic progenitors (HSC: hematopoietic stem cell, MPP: multipotent progenitors, GMP: Granulocyte-macrophage progenitors, CMP: common myeloid progenitors, MEP: megakaryocyte-erythroid progenitors) in BRAFwtScl+ and BRAFV600EScl+ animals. b, Flow cytometry gating strategy for bone marrow myeloid cells in BRAFwtScl+ and BRAFV600EScl+ animals. c, Experimental design of the CFU assays performed in Fig. 2e is shown.
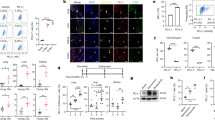
Extended Data Fig. 3 Senescence program in different LCH models.
a, Lineage negative BM cells isolated from either BRAFV600EScl+ CD45.2+ or BRAFwtScl+ CD45.2+ mice were injected intravenously together with lineage negative CD45.1+ BM cells into lethally irradiated CD45.1+ mice at a 2:1 ratio (CD45.2+: CD45.1+). Graph shows the ratio of CD45.2+ / CD45.1+ in blood circulation in each group measured 8 weeks after transplantation using flow cytometry (n = 8). b, Forward scatter measurement of lineage negative RosaYFP+ cells in the BM from BRAFV600EScl+ mice and BRAFwtScl+ control littermates (n = 5–7 mice per group). c, Expansion of human CD34+ cord blood HPC transduced with NGFR (NGFRhu) or BRAFV600E (BRAFV600Ehu) lentiviral constructs and cultured in stem-cell media. Data are representative of at least 3 experiments (n = 3 donors). d, Proliferation capacity of NGFRhu and BRAFV600Ehu HPC analyzed by Cell Trace Violet dilution, 92 hours after staining. e, Forward scatter measurement of NGFRhu and BRAFV600Ehu HPC (n = 3 different donors) f, Heat map representation of SASP genes analyzed by microarray expression profiling of BRAFV600E+ GFP+ HPC and NGFR+ GFP+ HPC 7 days after transduction (n = 4 different donors). g-h, Frozen or FFPE tissue sections isolated from humanized mice reconstituted with BRAFV600Ehu and NGFRhu HPC and stained for g, SAβGal activity or (h) p16INK4a. Graph shows the number of positive cells per mm2. Data are represented as mean ± s.e.m; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001 (unpaired two-sided t-tests, two-way ANOVA test for Extended Data Fig. 2c).
Extended Data Fig. 4 Human LCH lesions show features of senescence.
a, Heat map representation of SASP associated gene expression measured by RNA sequencing in purified CD34+ BM cells isolated from three LCH patients and one age-matched healthy donor (performed as duplicates). b, Skin from LCH patient and healthy age-matched donor were stained for CD207 and p16INK4a by immunohistochemistry. c, SAβGal activity of human tissues and LCH lesions. One representative image of healthy skin, naevus and LCH lesion are shown. Graph shows the number of SAβGal positive cells per mm2 in healthy skins (n = 3), naevus (n = 1) and LCH lesions (n = 3). d, Autopsy brain sections from a patient with neurodegenerative-LCH and circulating BRAFV600E+ peripheral blood cells. The H&E image shows a temporal lobe white matter injury with increased numbers of infiltrating plump cells. The immunohistochemical characterization of the enlarged plump cells shows a positivity for CD163, CD33, CD14, BRAFVE1 and p16INK4a. (*indicates perivascular space). e, Control tissue with rare p16INK4a positivity by immunohistochemistry in meningioma and no appreciable staining in human brain (non-diseased from autopsy). f, Brain section from human patient with neurodegenerative-LCH (see Fig. 4g) dual-stained for P2RY12 (green) and CD163 (red). Spindled P2RY12+ microglial cells surround the aggregates of plump activated CD163+ macrophages. There is no dual staining. There is diminished P2RY12 staining within the CD163+ macrophage rich aggregate. Image courtesy of Dr. Julia Kofler, MD UPMC Division of Neuropathology. Data are represented as mean ± s.e.m; statistical significance analyzed by an unpaired two-sided t-test is indicated by *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Extended Data Fig. 5 Rapamycin and senolytic treatments improve LCH outcome.
a, Human CD34+ cord blood HPC were transduced with BRAFV600E (BRAFV600Ehu) or NGFR control (NGFRhu) lentiviral vectors, cultured in stem-cell media and analyzed for gene expression of PI3K pathway genes using microarray expression profiling 7 days after transduction (n = 4 donors); b-c, BRAFV600EScl+ mice and BRAFwtScl+ control littermates were treated with rapamycin (0.5 mg/ kg body weight/ day) or DMSO for 10 days. b, Cartoon shows the experimental design to test the clinical benefit of rapamycin on LCH in mice. b, Graph shows the percentage of RosaYFP+ cells among CD45+ BM cells (n = 3–4 mice per group). d, BRAFV600EScl+ ATTAC+ mice and BRAFwtScl+ ATTAC+ control littermates were treated by tamoxifen for five days. Following tamoxifen injections, BRAFV600EScl+ ATTAC+ mice and BRAFwtScl+ ATTAC+ control littermates were treated by AP or vehicle during three weeks and sacrificed 4 weeks after tamoxifen injections. Representative flow plot showing the percentage of GFP+ cells among CD45+ live cells in the bone marrow from BRAFV600EScl+ ATTAC+ mice and BRAFwtScl+ ATTAC+ control littermates. e-h, BRAFV600EScl+ mice and BRAFwtScl+ control littermates were treated with ABT-263 (50 mg/ kg body weight/ day) or diluent for 21 days. e, Cartoon shows the experimental design to test the clinical benefit of ABT-263 on LCH in mice. f, Graph shows the absolute numbers of bone marrow RosaYFP+ cells (n = 2–3) g, Spleen weight and (h) lung infiltrating CD207+ cells isolated from mice treated with ABT-263 or diluent (n = 3–4) with representative images of lung tissue sections isolated from mice treated with ABT-263 or diluent and stained with CD207. Data are representative of 2 experiments (n = 3–4). Data are represented as mean ± s.e.m; statistical significance analyzed by an unpaired two-sided t-test is indicated by *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Supplementary information
Supplementary Table 1
Clinical characteristics of patients with LCH selected for the RNA-seq analysis of CD34+ BM hematopoietic progenitors.
Supplementary Table 2
Clinical characteristics of patients with LCH selected for the study.
Supplementary Table 3
Human CD34+ cord blood HPCs transduced with BRAFV600E or control NGFR lentiviral vector were cultured for 7 d in stem cell medium and analyzed using microarray sequencing.
Supplementary Table 4
RNA-seq data obtained from CD34+ cells purified from the BM of patients with LCH and healthy donors.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Rights and permissions
About this article
Cite this article
Bigenwald, C., Le Berichel, J., Wilk, C.M. et al. BRAFV600E-induced senescence drives Langerhans cell histiocytosis pathophysiology. Nat Med 27, 851–861 (2021). https://doi.org/10.1038/s41591-021-01304-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-021-01304-x
This article is cited by
-
Vemurafenib combined with chemotherapy achieved sustained remission in pediatric LCH: a multi-center observational study
Journal of Cancer Research and Clinical Oncology (2024)
-
BRAFV600E promotes DC3/monocyte differentiation in human gene-engineered HSPCs and causes multisystem histiocytosis
Leukemia (2023)
-
Signaling pathways, microenvironment, and targeted treatments in Langerhans cell histiocytosis
Cell Communication and Signaling (2022)
-
Molecular dissection on inhibition of Ras-induced cellular senescence by small t antigen of SV40
Cellular and Molecular Life Sciences (2022)
-
Histiocytic disorders
Nature Reviews Disease Primers (2021)