Abstract
Congenital hydrocephalus (CH), characterized by enlarged brain ventricles, is considered a disease of excessive cerebrospinal fluid (CSF) accumulation and thereby treated with neurosurgical CSF diversion with high morbidity and failure rates. The poor neurodevelopmental outcomes and persistence of ventriculomegaly in some post-surgical patients highlight our limited knowledge of disease mechanisms. Through whole-exome sequencing of 381 patients (232 trios) with sporadic, neurosurgically treated CH, we found that damaging de novo mutations account for >17% of cases, with five different genes exhibiting a significant de novo mutation burden. In all, rare, damaging mutations with large effect contributed to ~22% of sporadic CH cases. Multiple CH genes are key regulators of neural stem cell biology and converge in human transcriptional networks and cell types pertinent for fetal neuro-gliogenesis. These data implicate genetic disruption of early brain development, not impaired CSF dynamics, as the primary pathomechanism of a significant number of patients with sporadic CH.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The sequencing data for all CH parent–offspring trios and singletons reported in this study have been deposited in the NCBI Database of Genotypes and Phenotypes under accession number phs000744.v4.p2. Our in-house R and Python pipelines and codes are available upon request.
Code availability
Our in-house Python and R pipelines are available from the corresponding author on request.
References
Albright, A. L., Adelson, P. D. & Pollack, I. F. Principles and Practice of Pediatric Neurosurgery (Thieme, 2008).
Bondurant, C. P. & Jimenez, D. F. Epidemiology of cerebrospinal fluid shunting. Pediatr. Neurosurg. 23, 254–258 (1995).
Tully, H. M. & Dobyns, W. B. Infantile hydrocephalus: a review of epidemiology, classification and causes. Eur. J. Med. Genet 57, 359–368 (2014).
Lindquist, B., Carlsson, G., Persson, E. K. & Uvebrant, P. Behavioural problems and autism in children with hydrocephalus: a population-based study. Eur. Child Adolesc. Psychiatry 15, 214–219 (2006).
Kahle, K. T., Kulkarni, A. V., Limbrick, D. D. Jr. & Warf, B. C. Hydrocephalus in children. Lancet 387, 788–799 (2016).
Chervenak, F. A. et al. Outcome of fetal ventriculomegaly. Lancet 2, 179–181 (1984).
Haverkamp, F. et al. Congenital hydrocephalus internus and aqueduct stenosis: aetiology and implications for genetic counselling. Eur. J. Pediatrics 158, 474–478 (1999).
Kousi, M. & Katsanis, N. The genetic basis of hydrocephalus. Annu Rev. Neurosci. 39, 409–435 (2016).
Furey, C. G. et al. De novo mutation in genes regulating neural stem cell fate in human congenital hydrocephalus. Neuron 99, 302–314 e304 (2018).
Duran, D. et al. Mutations in chromatin modifier and ephrin signaling genes in vein of Galen malformation. Neuron 101, 429–443 (2019).
Duy, P. Q., Furey, C. G. & Kahle, K. T. Trim71/lin-41 links an ancient miRNA pathway to human congenital hydrocephalus. Trends Mol. Med. 25, 467–469 (2019).
Welte, T. et al. The RNA hairpin binder TRIM71 modulates alternative splicing by repressing MBNL1. Genes Dev. 33, 1221–1235 (2019).
Narayanan, R. et al. Loss of BAF (mSWI/SNF) complexes causes global transcriptional and chromatin state changes in forebrain development. Cell Rep. 13, 1842–1854 (2015).
Da, G. et al. Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes. Proc. Natl Acad. Sci. USA 103, 2057–2062 (2006).
Harmacek, L. et al. A unique missense allele of BAF155, a Core BAF chromatin remodeling complex protein, causes neural tube closure defects in mice. Developmental Neurobiol. 74, 483–497 (2014).
Liu, P., Cheng, H., Roberts, T. M. & Zhao, J. J. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Disco. 8, 627–644 (2009).
Li, L., Liu, F. & Ross, A. H. PTEN regulation of neural development and CNS stem cells. J. Cell Biochem. 88, 24–28 (2003).
Chalhoub, N. & Baker, S. J. PTEN and the PI3-kinase pathway in cancer. Annu Rev. Pathol. 4, 127–150 (2009).
Keppler-Noreuil, K. M., Parker, V. E., Darling, T. N. & Martinez-Agosto, J. A. Somatic overgrowth disorders of the PI3K/AKT/mTOR pathway & therapeutic strategies. Am. J. Med. Genet. C. Semin. Med. Genet. 172, 402–421 (2016).
Riviere, J. B. et al. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat. Genet. 44, 934–940 (2012).
Oda, K. et al. PIK3CA cooperates with other phosphatidylinositol 3′-kinase pathway mutations to effect oncogenic transformation. Cancer Res. 68, 8127–8136 (2008).
Dogruluk, T. et al. Identification of variant-specific functions of PIK3CA by rapid phenotyping of rare mutations. Cancer Res. 75, 5341–5354 (2015).
Foerster, P. et al. mTORC1 signaling and primary cilia are required for brain ventricle morphogenesis. Development 144, 201–210 (2017).
Martinez-Glez, V. et al. Macrocephaly-capillary malformation: analysis of 13 patients and review of the diagnostic criteria. Am. J. Med. Genet. A 152A, 3101–3106 (2010).
O’Rourke, D. J., Twomey, E., Lynch, S. A. & King, M. D. Cortical dysplasia associated with the PTEN mutation in Bannayan–Riley–Ruvalcaba syndrome: a rare finding. Clin. Dysmorphol. 21, 91–92 (2012).
Chen, H. H. et al. Immune dysregulation in patients with PTEN hamartoma tumor syndrome: analysis of FOXP3 regulatory T cells. J. Allergy Clin. Immunol. 139, 607–620 (2017).
Sarquis, M. S. et al. Distinct expression profiles for PTEN transcript and its splice variants in Cowden syndrome and Bannayan–Riley–Ruvalcaba syndrome. Am. J. Hum. Genet. 79, 23–30 (2006).
Groszer, M. et al. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science 294, 2186–2189 (2001).
Pilarski, R. & Eng, C. Will the real Cowden syndrome please stand up (again)? Expanding mutational and clinical spectra of the PTEN hamartoma tumour syndrome. J. Med. Genet. 41, 323–326 (2004).
Mirzaa, G. M. et al. Association of MTOR mutations with developmental brain disorders, including megalencephaly, focal cortical dysplasia, and pigmentary mosaicism. JAMA Neurol. 73, 836–845 (2016).
Baynam, G. et al. A germline MTOR mutation in aboriginal Australian siblings with intellectual disability, dysmorphism, macrocephaly, and small thoraces. Am. J. Med. Genet. A 167, 1659–1667 (2015).
Jacquet, B. V. et al. FoxJ1-dependent gene expression is required for differentiation of radial glia into ependymal cells and a subset of astrocytes in the postnatal brain. Development 136, 4021–4031 (2009).
Divina, P., Kvitkovicova, A., Buratti, E. & Vorechovsky, I. Ab initio prediction of mutation-induced cryptic splice-site activation and exon skipping. Eur. J. Hum. Genet. 17, 759–765 (2009).
Schonichen, A. & Geyer, M. Fifteen formins for an actin filament: a molecular view on the regulation of human formins. Biochim. Biophys. Acta 1803, 152–163 (2010).
Lian, G., Chenn, A., Ekuta, V., Kanaujia, S. & Sheen, V. Formin 2 regulates lysosomal degradation of wnt-associated β-catenin in neural progenitors. Cereb. Cortex 29, 1938–1952 (2019).
Lian, G. et al. Filamin A- and formin 2-dependent endocytosis regulates proliferation via the canonical Wnt pathway. Development 143, 4509–4520 (2016).
Gavino, C. & Richard, S. Patched1 haploinsufficiency impairs ependymal cilia function of the quaking viable mice, leading to fatal hydrocephalus. Mol. Cell. Neurosci. 47, 100–107 (2011).
Palma, V. et al. Sonic hedgehog controls stem cell behavior in the postnatal and adult brain. Dev. (Camb., Engl.) 132, 335–344 (2005).
Palma, V. & Ruiz i Altaba, A. Hedgehog-GLI signaling regulates the behavior of cells with stem cell properties in the developing neocortex. Development 131, 337–345 (2004).
Bult, C. J. et al. Mouse genome database (MGD) 2019. Nucleic Acids Res. 47, D801–D806 (2019).
Hehr, U. et al. Novel POMGnT1 mutations define broader phenotypic spectrum of muscle-eye-brain disease. Neurogenetics 8, 279–288 (2007).
Manzini, M. C. et al. Exome sequencing and functional validation in zebrafish identify GTDC2 mutations as a cause of Walker-Warburg syndrome. Am. J. Hum. Genet. 91, 541–547 (2012).
Walker, R. L. et al. Genetic control of expression and splicing in developing human brain informs disease mechanisms. Cell 179, 750–771 (2019).
Polioudakis, D. et al. A single-cell transcriptomic atlas of human neocortical development during mid-gestation. Neuron 103, 785–801 (2019).
Kurata, H. et al. Neurodevelopmental disorders in children with macrocephaly: a prevalence study and PTEN gene analysis. Brain Dev. 40, 36–41 (2018).
Palmen, S. J. et al. Increased gray-matter volume in medication-naive high-functioning children with autism spectrum disorder. Psychol. Med. 35, 561–570 (2005).
Gilmore, J. H. et al. Outcome in children with fetal mild ventriculomegaly: a case series. Schizophrenia Res. 48, 219–226 (2001).
Jin, S. C. et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat. Genet. 49, 1593–1601 (2017).
Iossifov, I. et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 (2014).
Wallmeier, J. et al. De novo mutations in FOXJ1 result in a motile ciliopathy with hydrocephalus and randomization of left/right body asymmetry. Am. J. Hum. Genet. 105, 1030–1039 (2019).
Guerra, M. M. et al. Cell junction pathology of neural stem cells is associated with ventricular zone disruption, hydrocephalus, and abnormal neurogenesis. J. Neuropathol. Exp. Neurol. 74, 653–671 (2015).
Wagner, C. et al. Cellular mechanisms involved in the stenosis and obliteration of the cerebral aqueduct of hyh mutant mice developing congenital hydrocephalus. J. Neuropathol. Exp. Neurol. 62, 1019–1040 (2003).
Zega, K. et al. Dusp16 deficiency causes congenital obstructive hydrocephalus and brain overgrowth by expansion of the neural progenitor pool. Front Mol. Neurosci. 10, 372 (2017).
Henzi, R. et al. Neural stem cell therapy of foetal onset hydrocephalus using the HTx rat as experimental model. Cell Tissue Res. 381, 141–161 (2020).
McAllister, J. P. et al. Ventricular zone disruption in human neonates with intraventricular hemorrhage. J. Neuropathol. Exp. Neurol. 76, 358–375 (2017).
Movsas, T. Z. et al. Autism spectrum disorder is associated with ventricular enlargement in a low birth weight population. J. Pediatrics 163, 73–78 (2013).
Li, M. et al. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 362, eaat7615 (2018).
Etchegaray, A., Juarez-Penalva, S., Petracchi, F. & Igarzabal, L. Prenatal genetic considerations in congenital ventriculomegaly and hydrocephalus. Childs Nerv. Syst. 36, 1645–1660 (2020).
Raudvere, U. et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198 (2019).
Krumm, N. et al. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 47, 582–588 (2015).
McKenna, A. et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Van der Auwera, G. A. et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinforma. 43, 11–33 (2013). 11 10.
Garrison E. M. G. Haplotype-based variant detection from short-read sequencing. Preprint at arXiv https://arxiv.org/abs/1207.3907 (2012).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Taliun, D. et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Preprint at bioRxiv https://doi.org/10.1101/563866 (2019).
Samocha, K. et al. Regional missense constraint improves variant deleteriousness prediction. Preprint at bioRxiv https://doi.org/10.1101/148353 (2017).
Dong, C. et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 24, 2125–2137 (2015).
Wei, Q. et al. A Bayesian framework for de novo mutation calling in parents-offspring trios. Bioinformatics 31, 1375–1381 (2015).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Samocha, K. E. et al. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 46, 944–950 (2014).
Ware, J. S., Samocha, K. E., Homsy, J. & Daly, M. J. Interpreting de novo variation in human disease using denovolyzeR. Curr. Protoc. Hum. Genet. 87, 21–15 (2015).
Wang, M., Marco, P., Capra, V. & Kibar, Z. Update on the role of the non-canonical wnt/planar cell polarity pathway in neural tube defects. Cells 8, 1198 (2019).
Tissir, F. & Goffinet, A. M. Shaping the nervous system: role of the core planar cell polarity genes. Nat. Rev. Neurosci. 14, 525–535 (2013).
Shaheen, R. et al. The genetic landscape of familial congenital hydrocephalus. Ann. Neurol. 81, 890–897 (2017).
Ruzzo, E. K. et al. Inherited and de novo genetic risk for autism impacts shared networks. Cell 178, 850–866 (2019).
Satterstrom, F. K. et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 180, 568–584 (2020).
Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature 542, 433–438 (2017).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinforma. 9, 559 (2008).
Li, J. et al. Application of weighted gene co-expression network analysis for data from paired design. Sci. Rep. 8, 622 (2018).
Acknowledgements
We are grateful to the patients and their families who participated in this research. We thank the Hydrocephalus Association (HA) for their support. We also thank J. Koschnitzky (HA), J. Rockefeller (Yale), J. Freeman (Yale) and J. Nicolleli (Yale) for their help and support. This work is supported by the Yale–National Institutes of Health (NIH) Center for Mendelian Genomics (5U54HG006504); NIH Director’s Pioneer Award DP1HD086071 and NIH Director’s Transformative Award 1R01AI145057 (S.J.S.); R01 NS111029-01A1, R01 NS109358, K12 228168 and the Rudi Schulte Research Institute (K.K.); NIH Medical Scientist Training Program (NIH/National Institute of General Medical Sciences Grant T32GM007205); NIH Clinical and Translational Science Award from the National Center for Advancing Translational Science (TL1 TR001864); James Hudson Brown – Alexander B. Coxe Fellowship at Yale School of Medicine, the American Heart Association Postdoctoral Fellowship (18POST34060008), the K99/R00 Pathway to Independence Award (K99HL143036 and R00HL143036-02) (S.C.J.); the American Heart Association Predoctoral Fellowship (19PRE34380842, W.D.); the Pediatric Hydrocephalus Foundation (P.H.F.). We thank M. C. Kruer at Phoenix Children’s Hospital and H. Zhao at Yale School of Public Health for critical discussion.
Author information
Authors and Affiliations
Contributions
S.C.J., R.P.L. and K.T.K. contributed to study design and conceptualization. C.G.F., A.T.T., C.N.-W., S.P., A.A.A., H.S., A.D., S.C., W.S., P.Q.D., T.D., B.C.R., A.M., J.R.B., E.M.K., P.S., C.H., B.K., S.J.S., M.L.J.A., E.J.H., L.R.M., J.K.K., J.G., F.T.M., A.J.K., W.E.B., E.R.S., B.C.W., D.D.L., G.H., E.M.J., B.J.I., J.M.J., K.B., S.M., C.C., S.L.A., B.G., Y.B., Y.S., C.C.D., M.L.D., M.G., R.P.L. and K.T.K. provided cohort ascertainment, recruitment and phenotypic characterization. I.R.T., C.C., K.B. and S.M. performed WES production and validation. S.C.J., W.D., X.Z., C.G.F., J.R.K. and M.C.S. conducted WES analysis. S.C.J., W.D., S.P., R.L.W., L.G., B.L. and Q.L. performed statistical analysis. C.N.-W. performed Sanger sequencing validation. A.J.K. and A.M.-D.-L. performed neuroimaging characterization. S.H. and H.P.P. conducted biophysical simulation. C.N.-W., K.B., S.M., S.L.A., N.S., D.H.G., M.G., R.P.L. and K.T.K. provided resources. S.C.J., A.J.K., S.P., W.D., S.L.A., R.P.L. and K.T.K. wrote and reviewed manuscript. S.C.J., C.N.-W., R.P.L. and K.T.K. administered the project. R.P.L. and K.T.K. acquired funding and supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Kate Gao and Jerome Staal were the primary editors on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
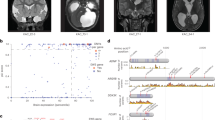
Extended Data Fig. 1 De novo, transmitted, and unphased mutations in TRIM71.
a, Pedigrees and sequencing electropherograms of Sanger sequencing depict all TRIM71 mutations in genomic DNA from CH probands. b, Representative T1 or T2-weighted brain magnetic resonance images for all available probands.
Extended Data Fig. 2 De novo, transmitted, and unphased mutations in SMARCC1.
a, Pedigrees and sequencing electropherograms of Sanger sequencing depict all SMARCC1 mutations in genomic DNA from CH probands. b, Representative T1 or T2-weighted brain magnetic resonance images or head CTs for all available probands.
Extended Data Fig. 3 De novo, transmitted, and unphased mutations in PIK3CA.
a, Pedigrees and sequencing electropherograms of Sanger sequencing depict all PIK3CA mutations in genomic DNA from CH probands. b, Representative T1 or T2-weighted brain magnetic resonance images for all available probands.
Extended Data Fig. 4 De novo, transmitted, and unphased mutations in PTEN.
a, Pedigrees and sequencing electropherograms of Sanger sequencing depict all PTEN mutations in genomic DNA from CH probands. b, Representative T1 or T2-weighted brain magnetic resonance images or head CTs for all available probands.
Extended Data Fig. 5 De novo, transmitted, and unphased mutations in MTOR.
a, Pedigrees and sequencing electropherograms of Sanger sequencing depict all MTOR mutations in genomic DNA from CH probands. b, Representative T1 or T2-weighted brain magnetic resonance images or head CTs for all available probands.
Extended Data Fig. 6 De novo and transmitted mutations in FOXJ1.
a, Pedigrees and sequencing electropherograms of Sanger sequencing depict all FOXJ1 mutations in genomic DNA from CH probands. b, Representative T1 or T2-weighted brain magnetic resonance images for all available probands.
Extended Data Fig. 7 De novo and transmitted mutations in FMN2.
a, Pedigrees and sequencing electropherograms of Sanger sequencing depict all FMN2 mutations in genomic DNA from CH probands. b, Representative T1 or T2-weighted brain magnetic resonance images for all available probands. c, The CRYP-SKIP algorithm prediction on splicing defects for FMN2: c.2137-2 A > G.
Extended Data Fig. 8 De novo, transmitted, and unphased mutations in PTCH1.
a, Pedigrees and sequencing electropherograms of Sanger sequencing depict all PTCH1 mutations in genomic DNA from CH probands. b, Representative T1 or T2-weighted brain magnetic resonance images or head CTs for all available probands.
Extended Data Fig. 9 Transmitted and unphased mutations in FXYD2.
a, Pedigrees and sequencing electropherograms of Sanger sequencing depict all FXYD2 mutations in genomic DNA from CH probands. b, Representative T1 or T2-weighted brain magnetic resonance images for all available probands. c, The CRYP-SKIP algorithm prediction on splicing defects for FXYD2: c.299-1 G > A. d, The CRYP-SKIP algorithm prediction on splicing defects for FXYD2: c.410 + 1 G > A.
Extended Data Fig. 10 Damaging recessive genotypes in human dystroglycanopathy genes and homologs of mouse hydrocephalus genes.
Available clinical-neuroimaging phenotypes of CH probands with damaging recessive mutations.
Supplementary information
Supplementary Information
Supplementary Note, Supplementary Figs. 1–9 and Supplementary References
Rights and permissions
About this article
Cite this article
Jin, S.C., Dong, W., Kundishora, A.J. et al. Exome sequencing implicates genetic disruption of prenatal neuro-gliogenesis in sporadic congenital hydrocephalus. Nat Med 26, 1754–1765 (2020). https://doi.org/10.1038/s41591-020-1090-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-020-1090-2
This article is cited by
-
The genetic basis of hydrocephalus: genes, pathways, mechanisms, and global impact
Fluids and Barriers of the CNS (2024)
-
A Review of Cerebrospinal Fluid Circulation and the Pathogenesis of Congenital Hydrocephalus
Neurochemical Research (2024)
-
Congenital hydrocephalus: new Mendelian mutations and evidence for oligogenic inheritance
Human Genomics (2023)
-
Development of shunt valves used for treating hydrocephalus: comparison with endoscopy treatment
Child's Nervous System (2023)
-
Heterozygous FOXJ1 Mutations Cause Incomplete Ependymal Cell Differentiation and Communicating Hydrocephalus
Cellular and Molecular Neurobiology (2023)