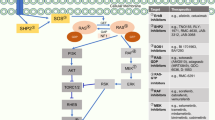
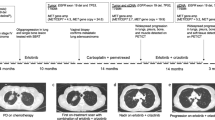
Abstract
MET exon 14 alterations are oncogenic drivers of non-small-cell lung cancers (NSCLCs)1. These alterations are associated with increased MET activity and preclinical sensitivity to MET inhibition2. Crizotinib is a multikinase inhibitor with potent activity against MET3. The antitumor activity and safety of crizotinib were assessed in 69 patients with advanced NSCLCs harboring MET exon 14 alterations. Objective response rate was 32% (95% confidence interval (CI), 21–45) among 65 response-evaluable patients. Objective responses were observed independent of the molecular heterogeneity that characterizes these cancers and did not vary by splice-site region and mutation type of the MET exon 14 alteration, concurrent increased MET copy number or the detection of a MET exon 14 alteration in circulating tumor DNA. The median duration of response was 9.1 months (95% CI, 6.4–12.7). The median progression-free survival was 7.3 months (95% CI, 5.4–9.1). MET exon 14 alteration defines a molecular subgroup of NSCLCs for which MET inhibition with crizotinib is active. These results address an unmet need for targeted therapy in people with lung cancers with MET exon 14 alterations and adds to an expanding list of genomically driven therapies for oncogenic subsets of NSCLC.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Upon request, and subject to certain criteria, conditions and exceptions (see https://www.pfizer.com/science/clinical-trials/trial-data-and-results for more information), Pfizer will provide access to individual de-identified patient data from Pfizer-sponsored global interventional clinical studies conducted for medicines, vaccines and medical devices (1) for indications that have been approved in the United States and/or European Union or (2) in programs that have been terminated (that is, development for all indications has been discontinued). Pfizer will also consider requests for the protocol, data dictionary and statistical analysis plan. Data may be requested from Pfizer trials 24 months after study completion. The de-identified patient data will be made available to researchers whose proposals meet the research criteria and other conditions, and for which an exception does not apply, via a secure portal. To gain access, data requestors must enter into a data access agreement with Pfizer.
References
Kong-Beltran, M. et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res. 66, 283–289 (2006).
Ma, P. C. et al. Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non-small cell lung cancer. Cancer Res. 65, 1479–1488 (2005).
Cui, J. J. et al. Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK). J. Med. Chem. 54, 6342–6363 (2011).
Awad, M. M. et al. MET exon 14 mutations in non-small-cell lung cancer are associated with advanced age and stage-dependent MET genomic amplification and c-met overexpression. J. Clin. Oncol. 34, 721–730 (2016).
Tong, J. H. et al. MET amplification and exon 14 splice site mutation define unique molecular subgroups of non-small cell lung carcinoma with poor prognosis. Clin. Cancer Res. 22, 3048–3056 (2016).
Schrock, A. B. et al. Characterization of 298 patients with lung cancer harboring MET exon 14 skipping alterations. J. Thorac. Oncol. 11, 1493–1502 (2016).
Frampton, G. M. et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 5, 850–859 (2015).
Drilon, A., Cappuzzo, F., Ou, S. I. & Camidge, D. R. Targeting MET in lung cancer: Will expectations finally be MET? J. Thorac. Oncol. 12, 15–26 (2017).
Onozato, R. et al. Activation of MET by gene amplification or by splice mutations deleting the juxtamembrane domain in primary resected lung cancers. J. Thorac. Oncol. 4, 5–11 (2009).
Drilon, A. MET exon 14 alterations in lung cancer: exon skipping extends half-life. Clin. Cancer Res. 22, 2832–2834 (2016).
Peschard, P. et al. Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein. Mol. Cell 8, 995–1004 (2001).
Awad, M. M. Impaired c-Met receptor degradation mediated by MET exon 14 mutations in non-small-cell lung cancer. J. Clin. Oncol. 34, 879–881 (2016).
Liu, X. et al. Next-generation sequencing of pulmonary sarcomatoid carcinoma reveals high frequency of actionable MET gene mutations. J. Clin. Oncol. 34, 794–802 (2016).
Paik, P. K. et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov. 5, 842–849 (2015).
Heist, R. S. et al. MET exon 14 skipping in non-small cell lung cancer. Oncologist 21, 481–486 (2016).
Jenkins, R. W. et al. Response to crizotinib in a patient with lung adenocarcinoma harboring a MET splice site mutation. Clin. Lung Cancer 16, e101–e104 (2015).
Lee, C. et al. MET 14 deletion in sarcomatoid non-small-cell lung cancer detected by next-generation sequencing and successfully treated with a MET inhibitor. J. Thorac. Oncol. 10, e113–e114 (2015).
Mendenhall, M. A. & Goldman, J. W. MET-mutated NSCLC with major response to crizotinib. J. Thorac. Oncol. 10, e33–e34 (2015).
Awad, M. M. et al. Impact of MET inhibitors on survival among patients with non-small cell lung cancer harboring MET exon 14 mutations: a retrospective analysis. Lung Cancer 133, 96–102 (2019).
Landi, L. et al. Crizotinib in MET deregulated or ROS1 rearranged pretreated non-small-cell lung cancer (METROS): a phase II, prospective, multicentre, two-arms trial. Clin. Cancer Res. https://doi.org/10.1158/1078-0432.CCR-19-0994 (2019).
Noonan, S. A. et al. Identifying the appropriate FISH criteria for defining MET copy number-driven lung adenocarcinoma through oncogene overlap analysis. J. Thorac. Oncol. 11, 1293–1304 (2016).
Genovese, G. et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371, 2477–2487 (2014).
Camidge, D. R. et al. Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: updated results from a phase 1 study. Lancet Oncol. 13, 1011–1019 (2012).
Shaw, A. T. et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N. Engl. J. Med. 371, 1963–1971 (2014).
Shaw, A. T. et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N. Engl. J. Med. 368, 2385–2394 (2013).
Benayed, R. et al. High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no mitogenic driver alteration detected by dna sequencing and low tumor mutation burden. Clin. Cancer Res. 25, 4712–4722 (2019).
Drilon, A. et al. Efficacy and safety of crizotinib in patients with advanced MET exon 14-altered non-small cell lung cancer (NSCLC). J. Clin. Oncol. 34, https://doi.org/10.1200/JCO.2016.34.15_suppl.108 (2016).
Shepherd, F. A. et al. Prospective randomized trial of docetaxel versus best supportive care in patients with non-small-cell lung cancer previously treated with platinum-based chemotherapy. J. Clin. Oncol. 18, 2095–2103 (2000).
Garon, E. B. et al. Ramucirumab plus docetaxel versus placebo plus docetaxel for second-line treatment of stage IV non-small-cell lung cancer after disease progression on platinum-based therapy (REVEL): a multicentre, double-blind, randomised phase 3 trial. Lancet 384, 665–673 (2014).
Scagliotti, G. V. et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J. Clin. Oncol. 26, 3543–3551 (2008).
Sandler, A. et al. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N. Engl. J. Med. 355, 2542–2550 (2006).
Socinski, M. A. et al. Atezolizumab for first-line treatment of metastatic nonsquamous NSCLC. N. Engl. J. Med. 378, 2288–2301 (2018).
National Comprehensive Cancer Network. NCCN clinical practice guidelines: Non-Small Cell Lung Cancer (Version 6.2019) (2019).
Soria, J. C. et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N. Engl. J. Med. 378, 113–125 (2018).
Camidge, D. R. et al. Brigatinib versus crizotinib in ALK-positive non-small-cell lung cancer. N. Engl. J. Med. 379, 2027–2039 (2018).
Lee, G. D. et al. MET exon 14 skipping mutations in lung adenocarcinoma: Clinicopathologic implications and prognostic values. J. Thorac. Oncol. 12, 1233–1246 (2017).
Guo, R. et al. MET inhibitor resistance in patients with MET exon 14-altered lung cancers. J. Clin. Oncol. 37, https://doi.org/10.1200/JCO.2019.37.15_suppl.9006 (2019).
Wolf, J. et al. Capmatinib (INC280) in METΔex14-mutated advanced non-small cell lung cancer (NSCLC): Efficacy data from the phase II GEOMETRY mono-1 study. J. Clin. Oncol. 37, https://doi.org/10.1200/JCO.2019.37.15_suppl.9004 (2019).
Paik, P. K. et al. Phase II study of tepotinib in NSCLC patients with METex14 mutations. J. Clin. Oncol. 37, https://doi.org/10.1200/JCO.2019.37.15_suppl.9005 (2019).
Lu, S. et al. Preliminary efficacy and safety results of savolitinib treating patients with pulmonary sarcomatoid carcinoma (PSC) and other types of non-small cell lung cancer (NSCLC) harboring MET exon 14 skipping mutations. Cancer Res. 79, https://doi.org/10.1158/1538-7445.AM2019-CT031 (2019).
Palma, N. A. et al. Frequency of MET amplification determined by comprehensive next-generation sequencing (NGS) in multiple solid tumors and implications for use of MET inhibitors. J. Clin. Oncol. 31, https://doi.org/10.1200/jco.2013.31.15_suppl.11068 (2013).
Oken, M. M. et al. Toxicity and response criteria of the Eastern Cooperative Oncology Group. Am. J. Clin. Oncol. 5, 649–655 (1982).
Therasse, P. et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J. Natl Cancer Inst. 92, 205–216 (2000).
Kwak, E. L. et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 363, 1693–1703 (2010).
Kwak, E. L. et al. Clinical activity observed in a phase I dose escalation trial of an oral c-met and ALK inhibitor, PF-02341066. J. Clin. Oncol. 27, https://doi.org/10.1200/jco.2009.27.15s.3509 (2009).
Acknowledgements
We thank the patients, their families and caregivers, and participating clinical sites and teams. For biomarker analyses, we acknowledge A.C. Donahue and C. Deshpande for their support. J.N. Raksin and J.G. Martins at inScience Communications, Springer Healthcare, provided medical writing support, funded by Pfizer Inc. This study was supported by Pfizer. A.D., G.J.R. and P.K.P. are supported by the National Institutes of Health award P30 CA008748.
Author information
Authors and Affiliations
Contributions
A.D. wrote the first draft of the paper and served as an investigator on the study. J.W.C., J.W., S-H.I.O., D.R.C., B.J.S., G.A.O., L.C.V., G.J.R., R.S.H, G.I.S, M.S., T.H., H.H. and P.K.P served as investigators on the study and wrote the paper. M.M.A. and K.D.W. wrote the paper. D.A.M., S.C.W., S.L. and T.U. analyzed the data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
A.D. reports receiving fees for consulting/advisory board roles at AstraZeneca, Bayer, BeiGene, Blueprint Medicines, Genentech/Roche, Helsinn, Ignyta, Loxo/Lilly, Pfizer, TP Therapeutics, BergenBio, Exelixis, Tyra, Verastem, MORE Health, Abbvie, GlaxoSmithKlein, Teva, PharmaMar, Taiho, Merck, Puma, Merus and Takeda/Ariad/Millenium. J.W.C. has nothing to disclose. J.W. reports receiving fees for speaker or advisory board roles from AstraZeneca, Biodesix, BioMarck Pharmaceuticals, Clovis Oncology, Eli Lilly, EMD Serono, Genentech and Inivata, and that his institution has received grant/research support from AstraZeneca, Astellas, Celgene, Merck, Novartis and Pfizer. S.-H.I.O. reports receiving fees for serving on speaker bureaus for AstraZeneca, Genentech and Takeda, fees for speaker or advisory board roles from AstraZeneca, Foundation Medicine, Novartis, Pfizer, Roche/Genentech and Takeda, honoraria from ARIAD/Takeda, AstraZeneca, Foundation Medicine, Genentech/Roche, Novartis, Pfizer and Roche Pharma AG, and that his institution has received grant/research support from ARIAD, Astellas Pharma, AstraZeneca, AstraZeneca/MedImmune, Chugai Pharma, Clovis Oncology, GlaxoSmithKline, Ignyta, Peregrine Pharmaceuticals, Pfizer and Roche Pharma AG. D.R.C. reports that his institution has received grant/research support from Pfizer. B.J.S. reports receiving fees for speaker or advisory board roles for AstraZeneca, Bristol-Myers Squibb, Merck Sharp & Dohme, Novartis, Pfizer and Roche/Genentech, honoraria from AstraZeneca and Bristol-Myers Squibb, that his institution has received grant/research support from Pfizer, that he has collected royalties and/or is an IP rights/patent holder with Veristrat (Biodesix), and that he has received other payment (travel, accommodations, expenses) from AstraZeneca, Bristol-Myers Squibb, Merck, Novartis and Roche. G.A.O. reports receiving fees for advisory board roles with Amgen, Genentech, Novartis, Pfizer and Takeda, and that his institution has received grant/research support from Astra Zeneca, BMS, Clovis, Genentech, Ignyta, Merck, Novartis and Pfizer. L.C.V. reports receiving fees for speaker or advisory board roles for Pfizer. G.J.R. reports receiving fees for speaker or advisory board roles from Genentech, and that his institution has received grant/research support from Ariad, GlaxoSmithKline, Infinity Pharmaceuticals, Millenium, Novartis, Pfizer and Roche/Genetech. R.S.H. reports receiving honoraria from Boehringer Ingelheim, and that her institution has received grant/research support from Abbvie, Agios, Celgene, Corvus, Daichii, Debiopharm, Genentech/Roche, Incyte, Millenium, Novartis and Peregrine. M.M.A. reports receiving fees for consulting/advisory board roles with Abbvie, ARIAD, AstraZeneca/MedImmune, Boehringer Ingelheim, Bristol-Myers Squibb, Clovis Oncology, Foundation Medicine, Genentech, Merck, Nektar, Novartis, Pfizer and Syndax, and that his institution has received grant/research support from Bristol-Myers Squibb. G.I.S. reports receiving fees for speaker or advisory board roles from G1 Therapeutics, Lilly, Pfizer, Roche and Vertex Pharmaceuticals, and that his institution has received grant/research support from Pfizer and Lilly. M.S. reports receiving honoraria from AstraZeneca, Boehringer Ingelheim, Bristol-Myers Squibb, Chugai Pharmaceutical, Eli Lilly Japan, Merck, Novartis, Ono Pharmaceutical, Pfizer Japan and Taiho Pharmaceutical, and that his institution has received grant/research support from AstraZeneca, Boehringer Ingelheim, Bristol-Myers Squibb, Chugai Pharmaceutical, Eli Lilly Japan, Merck, Novartis, Ono Pharmaceutical, Pfizer Japan and Taiho Pharmaceutical. T.H. reports receiving fees for consulting/advisory board roles from Novartis, and receiving honoraria from AstraZeneca, Boehringer Ingelheim, Bristol-Meyers Squibb, Chugai Pharmaceutical, Clovis Oncology, Eli Lilly, Nippon, Novartis, Ono Pharmaceutical, Pfizer and Taiho Pharmaceutical. H.H. reports receiving fees for consulting/advisory board roles from AstraZeneca, Boehringer Ingelheim, Chugai Pharma and Lilly. D.A.M. reports being an employee of, and owning stock in, Pfizer. S.C.W. reports being an employee of, and owning stock in, Pfizer. S.L. reports being an employee of, and owning stock in, Pfizer. T.U. reports being an employee of, and owning stock in, Pfizer. K.D.W. reports being an employee of, and owning stock in, Pfizer. P.K.P. has nothing to disclose. No other potential conflict of interest relevant to this article was reported.
Additional information
Peer review information Javier Carmona was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Patient evaluation groups.
Shown is a flow diagram summarizing study enrollment and patient evaluation groups in the MET-exon-14-altered advanced NSCLC expansion cohort of the ongoing PROFILE 1001 study as of 31 January 2018. Patients who received ≥1 dose of crizotinib were included in the safety population and analyses of PFS and OS. OS, overall survival; PFS, progression-free survival.
Extended Data Fig. 2 Best percent change in target lesions from baseline in MET exon 14-altered NSCLCs and MET exon 14 alterations (splice site region and type) by central and local testing.
A plot of the best response to crizotinib in 52 patients with MET-exon-14-altered NSCLCs is shown. The bars indicate the best percentage change in the sum of target tumor measurements from baseline. a–d, Below the plot, retrospective central testing results for the MET exon 14 alteration splice site region (a) and mutation type (c) are depicted in relation to best response. For comparison, prospective local testing results for region (b) and type (d) are also included. In rows a and b, the splice acceptor region includes alterations in the splice acceptor region, polypyrimidine tract and branching point. Cases classified as unknown include MET exon 14 alterations for which DNA-coding region information was not available, such as alterations detected using an RNA-based assay. White space indicates that no results were reported by the central (rows a, c) or local (rows b, d) assay, or that the reported results could not be analyzed for the biomarker of interest. Retrospective central testing confirmed the presence of a MET exon 14 alteration in 88% of the 40 patients with tumor tissue analyzed. Central testing confirmation was not obtained for five patients with MET-exon-14-altered NSCLC as determined by local testing methods. Of these five central testing-negative patients, four did not pass full quality-control metrics (mostly attributed to low tumor purity or tumor input issues) but were reportable. One patient was determined to have MET-exon-14-altered NSCLC by local testing and ROS1 rearranged NSCLC by central testing (as indicated by an asterisk).
Extended Data Fig. 3 Concurrent alterations in tumor.
Shown are concurrent alterations observed by retrospective molecular profiling of archival tumor tissue (FoundationOne CDx). Each patient with available data is represented by a column. The colored rectangles above each column represent the best objective response to crizotinib. Within a column, each gene of interest for which a concurrent alteration is present is represented by a colored rectangle corresponding to the alteration type. Concurrent genomic alterations (average number of alterations per patient was 4.3 (range, 0 to 12)) were identified in tumor tissue from 35 of 40 (88%) patients with analyzable samples. MDM2 amplification was detected in non-responders, but not observed in responders. No notable response differences were seen in relation to absence or presence of TP53 mutation. AMP, amplification. SNV, single-nucleotide variant.
Extended Data Fig. 4 Progression-free survival by MET exon 14 alteration detection in ctDNA.
The Kaplan–Meier curves for progression-free survival in patients treated with crizotinib are shown according to detection of MET exon 14 alterations in ctDNA by plasma profiling. Progression-free survival was defined as the time from the date of the first dose of crizotinib to objective disease progression or death from any cause. Hash marks on the survival curve indicate censoring of data. *P value from two-sided log-rank test comparing survival distributions among ctDNA-positive versus ctDNA-negative patients. †HR from Cox proportional hazards regression – assuming proportional hazards, a HR >1 indicates a greater risk of disease progression or death among ctDNA-positive versus ctDNA-negative patients. ctDNA, circulating tumor DNA; HR, hazard ratio; NE, not estimable; PFS, progression-free survival.
Extended Data Fig. 5 Concurrent Alterations in ctDNA.
Shown are concurrent alterations observed by retrospective molecular profiling of baseline plasma samples (PlasmaSELECTR64). Each patient with available data is represented by a column. The colored rectangles above each column represent the best objective response to crizotinib. Within a column, each gene of interest for which a concurrent alteration is present is represented by a colored rectangle corresponding to the alteration type. Concurrent genomic alterations (average number of alterations per patient was 2.36 (range, 0 to 8)) were identified in ctDNA from 25 of 36 (69%) patients with analyzable samples.
Supplementary information
Supplementary Information
Supplementary Tables 1–6, Clinical Protocol, Statistical Analysis Plan, and Supplementary Statistical Analysis Plan
Rights and permissions
About this article
Cite this article
Drilon, A., Clark, J.W., Weiss, J. et al. Antitumor activity of crizotinib in lung cancers harboring a MET exon 14 alteration. Nat Med 26, 47–51 (2020). https://doi.org/10.1038/s41591-019-0716-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-019-0716-8
This article is cited by
-
Oncogenic alterations in advanced NSCLC: a molecular super-highway
Biomarker Research (2024)
-
TWIST1 is a critical downstream target of the HGF/MET pathway and is required for MET driven acquired resistance in oncogene driven lung cancer
Oncogene (2024)
-
MET exon 14 skipping mutation drives cancer progression and recurrence via activation of SMAD2 signalling
British Journal of Cancer (2024)
-
Efficacy and safety analysis of immunotherapy in non-small cell lung cancer patients with MET alterations
Clinical and Translational Oncology (2024)
-
Rare molecular subtypes of lung cancer
Nature Reviews Clinical Oncology (2023)