Abstract
B-cell lymphoma extra large (BCL-XL) is a well-validated cancer target. However, the on-target and dose-limiting thrombocytopenia limits the use of BCL-XL inhibitors, such as ABT263, as safe and effective anticancer agents. To reduce the toxicity of ABT263, we converted it into DT2216, a BCL-XL proteolysis-targeting chimera (PROTAC), that targets BCL-XL to the Von Hippel-Lindau (VHL) E3 ligase for degradation. We found that DT2216 was more potent against various BCL-XL-dependent leukemia and cancer cells but considerably less toxic to platelets than ABT263 in vitro because VHL is poorly expressed in platelets. In vivo, DT2216 effectively inhibits the growth of several xenograft tumors as a single agent or in combination with other chemotherapeutic agents, without causing appreciable thrombocytopenia. These findings demonstrate the potential to use PROTAC technology to reduce on-target drug toxicities and rescue the therapeutic potential of previously undruggable targets. Furthermore, DT2216 may be developed as a safe first-in-class anticancer agent targeting BCL-XL.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The MS data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) through the PRIDE partner repository with the dataset identifiers PXD010878 and PXD015454. The raw immunoblot images and files from statistical analyses are supplied as source data. Other raw and analyzed data files are available from the corresponding author upon reasonable request. A data availability statement is also included in the reporting summary linked to this article.
References
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell. 144, 646–674 (2011).
Singh, R., Letai, A. & Sarosiek, K. Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat. Rev. Mol. Cell Biol. 20, 175–193 (2019).
Igney, F. H. & Krammer, P. H. Death and anti-death: tumour resistance to apoptosis. Nat. Rev. Cancer 2, 277–288 (2002).
Ashkenazi, A., Fairbrother, W. J., Leverson, J. D. & Souers, A. J. From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors. Nat. Rev. Drug Discov. 16, 273–284 (2017).
Adams, J. M. & Cory, S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene 26, 1324–1337 (2007).
Reed, J. C. Bcl-2-family proteins and hematologic malignancies: history and future prospects. Blood 111, 3322–3330 (2008).
Thomas, S. et al. Targeting the Bcl-2 family for cancer therapy. Expert Opin. Ther. Targets 17, 61–75 (2013).
Opfermann, J. T. Attacking cancer’s Achilles heel: antagonism of antiapoptotic BCL-2 family members. FEBS J. 283, 2661–2675 (2016).
Garner, T. P., Lopez, A., Reyna, D. E., Spitz, A. Z. & Gavathiotis, E. Progress in targeting the BCL-2 family of proteins. Curr. Opin. Chem. Biol. 39, 133–142 (2017).
Delbridge, A. R., Grabow, S., Strasser, A. & Vaux, D. L. Thirty years of BCL-2: translating cell death discoveries into novel cancer therapies. Nat. Rev. Cancer 16, 99–109 (2016).
Delbridge, A. R. & Strasser, A. The BCL-2 protein family, BH3-mimetics and cancer therapy. Cell Death Differ. 22, 1071–1080 (2015).
Oltersdorf, T. et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 435, 677–681 (2005).
Tse, C. et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 68, 3421–3428 (2008).
Souers, A. J. et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 19, 202–208 (2013).
Tao, Z. F. et al. Discovery of a potent and selective BCL-XL inhibitor with in vivo activity. ACS Med. Chem. Lett. 5, 1088–1093 (2014).
Kotschy, A. et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature 538, 477–482 (2016).
Deeks, E. D. Venetoclax: first global approval. Drugs 76, 979–987 (2016).
Roberts, A. W. et al. Targeting BCL2 with venetoclax in relapsed chronic lymphocytic leukemia. N. Engl. J. Med. 374, 311–322 (2016).
Mason, K. D. et al. Programmed anuclear cell death delimits platelet life span. Cell. 128, 1173–1186 (2007).
Schoenwaelder, S. M. et al. Bcl-xL-inhibitory BH3 mimetics can induce a transient thrombocytopathy that undermines the hemostatic function of platelets. Blood 118, 1663–1674 (2011).
Kaefer, A. et al. Mechanism-based pharmacokinetic/pharmacodynamic meta-analysis of navitoclax (ABT-263) induced thrombocytopenia. Cancer Chemother. Pharmacol. 74, 593–602 (2014).
Itchaki, G. & Brown, J. R. The potential of venetoclax (ABT-199) in chronic lymphocytic leukemia. Adv. Hematol. 7, 270–287 (2016).
Perini, G. F., Ribeiro, G. N., Neto, J. V. P., Campos, L. T. & Hamerschlak, N. BCL-2 as therapeutic target for hematological malignancies. J. Hematol. Oncol. 11, 65 (2018).
Leverson, J. D. et al. Exploiting selective BCL-2 family inhibitors to dissect cell survival dependencies and define improved strategies for cancer therapy. Sci. Transl. Med. 7, 279ra40 (2015).
Amundson, S. A. et al. An informatics approach identifying markers of chemosensitivity in human cancer cell lines. Cancer Res. 60, 6101–6110 (2000).
Vogler, M. Targeting BCL2-proteins for the treatment of solid tumours. J. Adv Med. 2014, 943648 (2014).
Lai, A. C. & Crews, C. M. Induced protein degradation: an emerging drug discovery paradigm. Nat. Rev. Drug Discov. 16, 101–114 (2017).
Runcie, A. C., Chan, K. H., Zengerle, M. & Ciulli, A. Chemical genetics approaches for selective intervention in epigenetics. Curr. Opin. Chem. Biol. 33, 186–194 (2016).
Deshaies, R. J. Protein degradation: prime time for PROTACs. Nat. Chem. Biol. 11, 634–635 (2015).
Churcher, I. Protac-induced protein degradation in drug discovery: breaking the rules or just making new ones? J. Med. Chem. 61, 444–452 (2018).
Ohoka, N., Shibata, N., Hattori, T. & Naito, M. Protein knockdown technology: application of ubiquitin ligase to cancer therapy. Curr. Cancer Drug Targets 16, 136–146 (2016).
Lu, J. et al. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem. Biol. 22, 755–763 (2015).
Bondeson, D. P. et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 11, 611–617 (2015).
Lai, A. C. et al. Modular PROTAC design for the degradation of oncogenic BCR-ABL. Angew. Chem. Int. Ed. 55, 807–810 (2016).
Raina, K. et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc. Natl Acad. Sci. USA 113, 7124–7129 (2016).
Saenz, D. T. et al. Novel BET protein proteolysis-targeting chimera exerts superior lethal activity than bromodomain inhibitor (BETi) against post-myeloproliferative neoplasm secondary (s) AML cells. Leukemia 31, 1951–1961 (2017).
Winter, G. E. et al. Drug development. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348, 1376–1381 (2015).
Huang, H. T. et al. A chemoproteomic approach to query the degradable kinome using a multi-kinase degrader. Cell Chem. Biol. 25, 88–99.e6 (2018).
Bray, P. F. et al. The complex transcriptional landscape of the anucleate human platelet. BMC Genomics 14, 1 (2013).
Kissopoulou, A., Jonasson, J., Lindahl, T. L. & Osman, A. Next generation sequencing analysis of human platelet PolyA+ mRNAs and rRNA-depleted total RNA. PLoS One 8, e81809 (2013).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013).
Vogler, M. et al. BCL2/BCL-X(L) inhibition induces apoptosis, disrupts cellular calcium homeostasis, and prevents platelet activation. Blood. 117, 7145–7154 (2011).
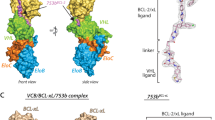
Gadd, M. S. et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol. 13, 514–521 (2017).
Nowak, R. P. et al. Plasticity in binding confers selectivity in ligand-induced protein degradation. Nat. Chem. Biol. 14, 706–714 (2018).
Riching, K. M. et al. Quantitative live-cell kinetic degradation and mechanistic profiling of PROTAC mode of action. ACS Chem. Biol. 13, 2758–2770 (2018).
Farmer, T., O’Neill, K. L., Naslavsky, N., Luo, X. & Caplan, S. Retromer facilitates the localization of Bcl-xL to the mitochondrial outer membrane. Mol. Biol. Cell 30, 1138–1146 (2019).
Smith, B. E. et al. Differential PROTAC substrate specificity dictated by orientation of recruited E3 ligase. Nat. Commun. 10, 131 (2019).
Morowski, M. et al. Only severe thrombocytopenia results in bleeding and defective thrombus formation in mice. Blood 121, 4938–4947 (2013).
Rinder, H. M. et al. Correlation of thrombosis with increased platelet turnover in thrombocytosis. Blood 91, 1288–1294 (1998).
Koch, R. et al. Biomarker-driven strategy for MCL1 inhibition in T-cell lymphomas. Blood 133, 566–575 (2018).
Berger, S. et al. Computationally designed high specificity inhibitors delineate the roles of BCL2 family proteins in cancer. eLife. 5, e20352 (2016).
Hikita, H. et al. Mcl-1 and Bcl-xL cooperatively maintain integrity of hepatocytes in developing and adult murine liver. Hepatology 50, 1217–1226.
Chen, J. et al. The Bcl-2/Bcl-X(L)/Bcl-w inhibitor, navitoclax, enhances the activity of chemotherapeutic agents in vitro and in vivo. Mol. Cancer Ther. 10, 2340–2349 (2011).
Ackler, S. et al. The Bcl-2 inhibitor ABT-263 enhances the response of multiple chemotherapeutic regimens in hematologic tumors in vivo. Cancer Chemother. Pharmacol. 66, 869–880 (2010).
Pompili, L., Porru, M., Caruso, C., Biroccio, A. & Leonetti, C. Patient-derived xenografts: a relevant preclinical model for drug development. J. Exp. Clin. Cancer Res. 35, 189 (2016).
Zengerle, M., Chan, K. H. & Ciulli, A. Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem. Biol. 10, 1770–1777 (2015).
Bondeson, D. P. et al. Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell Chem Biol. 25, 78–87.e5 (2018).
Lv, D.-W., Zhang, K. & Li, R. Interferon regulatory factor 8 regulates aspase-1 expression to facilitate Epstein–Barr virus reactivation in response to B cell receptor stimulation and chemical induction. PLoS Pathog. 14, e1006868 (2018).
Wiśniewski, J. R. et al. Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 (2009).
Nesvizhskii, A. I., Keller, A., Kolker, E. & Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 (2003).
Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote) omics data. Nat. Methods 13, 731–740 (2016).
Vizcaíno, J. A. et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 (2013).
US Food and Drug Administration. Bioanalytical Method Validation, Guidance for Industry (US Department of Health and Human Services Food and Drug Administration, 2018).
Acknowledgements
This study was supported by US National Institutes of Health (NIH) grants R01 CA211963 (D.Z.), R01 CA219836 (D.Z.), R01 GM109645 (R.H.), R01 CA205224 (R.H.), R01 CA200673 (W.Z.), R01 CA203834 (W.Z.), R21 CA223371 (G.Z.), R35 CA210065 (A.F.), R01 CA172809 (Y.-M.K.), Department of Defense grant BC180227 (W.Z.), the Schwab Charitable fund (M.K.) and CPRIT grant RP160716 (P.J.H.). Mass spectrometric support was provided by Alan J. Tackett and Samuel G. Mackintosh in the UAMS Proteomics Core Facility.
Author information
Authors and Affiliations
Contributions
S.K. designed, performed and analyzed most of the experiments and wrote the manuscript; X.Z. designed, synthesized and analyzed BCL-XL PROTACs and wrote the manuscript; D.L. designed, performed and analyzed the mass-spectrometry experiments, nanoBRET assay and wrote the manuscript; Y.H., D.T., J.S.W., J.P., W.Z., A.S., C.R.M, V.M.K., N.B., A. R. and G.H. performed and analyzed some experiments; P.Z and X.L. designed, synthesized and analyzed BCL-XL PROTACs; A.A.F. and Y.-M.K. provided the PDX T-ALL model and revised the manuscript; Q.Z. and M.K. designed and performed the PDX T-ALL study and analyzed and interpreted data and revised the manuscript; Y.Y., P.J.H, W.Z. and R.H. interpreted data and revised the manuscript; G.Z. designed and supervised the study, analyzed and interpreted data and revised the manuscript. D.Z. conceived, designed and supervised the study, analyzed and interpreted data, and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
S.K., X.Z, Y.H., P.Z., G.Z. and D.Z. are inventors of two pending patent applications for use of BCL-XL PROTACs as senolytic and antitumor agents. R.H., G.Z. and D.Z. are co-founders of and have equity in Dialectic Therapeutics, which develops BCL-XL PROTACs to treat cancer.
Additional information
Peer review information Javier Carmona was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
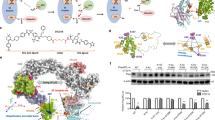
Extended Data Fig. 1 Expression of anti-apoptotic BCL-2 members and VHL in cancer, and the synthetic schemes of DT2216 and DT2216NC.
a, A representative of three immunoblot analyses of VHL in three different human tumor cell lines and platelets from three different individuals (indicated as units 1–3). b, Immunoblot analyses of the basal protein levels of BCL-XL, BCL-2, MCL-1 and VHL in different solid tumor cells. Data are representative of two independent experiments. c, BCL2L1 and VHL mRNA expression (log2 transformed) and mutational status were analyzed using TCGA PanCancer Atlas studies via cBioPortal. d, Synthetic schemes of DT2216, DT2216NC and VHL-L. Reagents and conditions: (i) (1) N-methylmorpholine, isobutyl chloroformate, THF, –25 °C then −15 °C; (2) NaBH4, H2O, –15 °C; (ii) Bu3P, diphenyl disulfide, toluene, 80 °C; (iii) DIBAL-H, toluene, –78 °C (iv) compound 5, NaBH(OAc)3, TEA, DCM; (v) TFA, DCM; (vi) compound 8, TEA, acetonitrile, reflux; (vii) compound 10, EDCI, DMAP, DCM; (viii) Zn, HOAc, THF; (ix) HATU, TEA, DCM; (x) (1) LiOH monohydrate, methanol, H2O; (2) compound 12, HATU, TEA, DCM; (xi) Ac2O, TEA, DCM.
Extended Data Fig. 2 The BCL-XL degradation by DT2216 is rapid and long lasting.
a, Immunoblot analysis of BCL-XL expression in MOLT-4 cells after they were treated with DT2216 for various durations as indicated. A representative immunoblot is presented on the top panel. Densitometric analysis of BCL-XL expression is presented on the bottom panel as mean of two independent experiments. Each symbol represents data (% of 0 h) from an individual experiment. b, Analysis of BCL-XL expression by immunoblot in MOLT-4 cells treated with DT2216 for 16 h followed by drug withdrawal and then cultured without DT2216 for 0 to 48 h as indicated. A representative immunoblot is on the top panel. Densitometric analysis of BCL-XL expression is presented on the bottom panel as the mean of two independent experiments. Each symbol represents data (% of Veh) from an individual experiment. c, Caspase-3 activity in MOLT-4 cells was measured 24 h after they were treated with 1 µM of DT2216 or ABT263. Data are presented as the mean (n = 2 technical replicates) of a representative experiment. Each symbol represents data (% of Veh) from an individual replicate. Similar results were obtained in an additional independent experiment. d, Representative immunoblot to confirm CRISPR–Cas9-mediated double knockout of BAX and BAK in H146 cells. The experiment was repeated independently one more time with similar results.
Extended Data Fig. 3 ABT263 or VHL-L blocks the formation of ternary complex of BCL-XL, DT2216 and VHL.
a, Recombinant His-tagged BCL-XL protein (100 nM) was incubated with the VHL-Elongin B/C complex (50 nM) with 0.039 µM of DT2216. The ternary complex formation was abrogated in the presence of ABT263 (1 µM) or VHL-L (10 µM). Data are expressed as mean (n = 2 technical replicates). Each symbol represents data from an individual replicate. b, The negative-control of DT2216 (DT2216NC) cannot form a ternary complex with BCL-XL and the VHL complex. Recombinant His-tagged BCL-XL protein (100 nM) was incubated with the VHL-Elongin B/C complex (50 nM) with increasing concentrations of DT2216 or DT2216NC. Data are expressed as mean (n = 2 technical replicates).
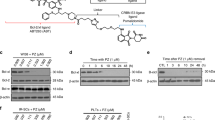
Extended Data Fig. 4 DT2216 binds to both BCL-XL and BCL-2 but cannot degrade BCL-2.
a, The binding affinities of DT2216 and ABT263 towards BCL-XL, BCL-2 and BCL-W were measured by AlphaScreen and are represented in terms of inhibition constant (Ki). The data are average of two independent experiments each performed in duplicates. b,c, Immunoblot analyses of BCL-2 and BCL-W are shown after the RS4 cells were treated with indicated concentrations of DT2216 for 16 h (top), or with 1 µM of DT2216 for indicated durations (bottom). d, Cell viability of BCL-2-dependent RS4 cells after treatment with increasing concentrations of DT2216, ABT199 or ABT263 for 72 h. Data are presented as mean ± s.d. from six replicate cell cultures in one experiment. Similar results were obtained in two additional independent experiments for DT2216. The EC50 of DT2216 is the average of three independent experiments. e, Cell viability of MCL-1-dependent H929 cells after treatment with increasing concentrations of DT2216 or S63845 for 72 h. Data are presented as mean ± s.d. from six replicate cell cultures in one experiment. f, Schematic representation of the proteomic assay shown in Fig. 3c. g, Representative immunoblot analyses are shown after the cells were treated with indicated concentrations of PZ-15227 for 16 h (top) or with 0.1 µM of PZ-15227 for indicated time points (bottom). Similar results were obtained in one more independent experiment. h, CETSA assay for BCL-XL and BCL-2. MOLT-4 (for BCL-XL) or RS4 (for BCL-2) cells were treated with DMSO, or 1 µM of DT2216 for 6 h. Raw band intensities are mean of two independent experiments.
Extended Data Fig. 5 Drug metabolism and pharmacokinetic/pharmacodynamic profile of DT2216.
a, DT2216 stability in mouse blood and plasma. b, DT2216 liver microsomal stability. c, Plasma concentrations of DT2216 after a single administration of 2 mg per kg body weight (i.v. injection), 20 mg per kg body weight (i.p. injection) or 20 mg per kg body weight (p.o. administration) are presented as mean ± s.d. (n = 3 mice per group). These studies were done by BioDuro, a global contract research organization, through a contract. d, MRM chromatograms of (A) DT2216 in drug-free brain homogenate, (B) internal standard in spiked drug-free tumor homogenate (40 ng ml–1), (C) DT2216 in spiked tumor homogenate (5 ng ml–1; LLOQ), (D) DT2216 in tumor sample taken from vehicle dosed mouse at 24 h and (E) DT2216 in tumor sample taken at 24 h after 15 mg per kg body weight, i.p. administration. e, Representative immunoblot analysis of BCL-XL in tumors at different durations after DT2216 (DT, 15 mg per kg body weight, i.p.) administration (n = 2 mice in vehicle and DT2216 groups at each time points). Similar results were obtained in two more immunoblot experiments.
Extended Data Fig. 6 DT2216 induces BCL-XL degradation and apoptosis in MOLT-4 T-ALL xenografts and suppresses their growth in a dose-dependent manner.
a,b, T-ALL xenograft tumors were collected 2 d after female CB-17 SCID mice received 1 i.p. injection of DT2216 at 7.5 mg per kg body weight and 15 mg per kg body weight. A single immunoblot analysis of BCL-XL, BCL-2, MCL-1, cleaved and full length caspase-3 and PARP1 in the tumors is shown. c, Changes in tumor volume over time after the start of treatment with vehicle (Veh), or DT2216 (7.5 or 15 mg per kg body weight, q7d, i.p.). All the data presented are mean ± s.e.m. (n = 8 mice in vehicle, 8 mice in 7.5 mg DT2216 per kg body weight and 7 mice in 15 mg DT2216 per kg body weight). The data from the 15 mg DT2216 per kg body weight group are also presented in Extended Data Fig. 8f, in which the tumor size in these mice were continuously monitored till day 55 post-treatment.
Extended Data Fig. 7 Images of MOLT-4-tumor-bearing mice and excised tumors.
The images shown (quantification data are shown in Fig. 4f,g) were captured at the end of the experiment, when the mice were treated with vehicle, DT2216 (15 mg per kg body weight, q7d, i.p.) or ABT263 (50 mg per kg body weight, qd, p.o.). The tumor-bearing mice (shown in the top) and harvested tumors (shown in the bottom) are not placed in an identical order.
Extended Data Fig. 8 DT2216 induces regression of MOLT-4 xenografts without causing thrombocytopenia.
a, Illustration of the experimental design of the MOLT-4 T-ALL xenograft mouse model. b, Blots that are representative of two independent immunoblot analyses of BCL-XL, cleaved and full-length caspase-3 and PARP1 in MOLT-4 T-ALL collected 2 d after tumor-bearing mice were treated with a single injection of vehicle or DT2216 (15 mg per kg body weight, i.p.). c, Blood platelets (PLT), white blood cells (WBC), lymphocytes (LYM) and granulocytes (GRA) were numerated 1 d after first treatment with vehicle, DT2216 (15 mg per kg body weight, i.p.) or ABT263 (15 mg per kg body weight, i.p.) as shown in a. a and b represent statistical significance versus Veh and DT2216, respectively, determined by one-way ANOVA and Tukey’s multiple-comparison test. d, Body-weight changes in mice after the start of treatment with vehicle, DT2216 or ABT263 as shown in a. e, Numeration of PLT 1 d after the sixth dose of DT2216 (15 mg per kg body weight, q7d, i.p. or 15 mg per kg body weight, q4d, i.p.). Data are presented as mean ± s.e.m. (n = 2 mice in vehicle, 7 mice in DT2216 q7d and 6 mice in DT2216 q4d). f, Changes in tumor volume over time after the start of treatment with vehicle, DT2216 or ABT263 as shown in a. Data presented in c, d and f are mean ± s.e.m. (n = 6 mice for vehicle group and 7 mice each for DT2216 and ABT263 groups). Each symbol in c and e represents data from an individual animal.
Extended Data Fig. 10 Anti-leukemic activity of DT2216 alone and in combination with chemotherapy in T-ALL PDX models.
a, Eight-week-old female NSG mice (n = 20 mice) were injected with 1 × 106 D115 cells 24 h after 0.25-Gy irradiation. Upon engraftment, mice were randomized to receive vehicle, DT2216 (15 mg per kg body weight, i.p., q4d for 3 weeks), chemotherapy (Chemo (vincristine 0.15 mg per kg body weight + dexamethasone 5 mg per kg body weight + l-asparaginase 1,000 units per kilogram body weight, i.p., q7d for 3 weeks)), or the combination of DT2216 with chemotherapy. Disease burden was followed by engraftment in bone marrow by checking hCD45% in bone marrow aspiration samples through flow cytometry. Data are presented as mean ± s.e.m. (n = 5 mice in each group). Each symbol represents data from an individual animal, and the middle horizontal line represents the mean. b, Mice survival was followed and statistical significance was determined by log-rank test (n = 5 mice in each group). c, Eight-week-old female NSG mice (n = 20 mice) were injected with 1 × 106 332X-luci cells 24 h after 0.25-Gy irradiation. Upon engraftment, mice were randomized to receive vehicle, DT2216, chemotherapy or the combination of DT2216 with chemotherapy as mentioned in a. Disease burden was followed by checking hCD45% in peripheral blood samples (retro orbital) through flow cytometry. Data are presented as mean ± s.e.m. (n = 5 mice in each group). Each symbol represents data from an individual animal, and the middle horizontal line represents the mean. d, Mice survival was followed, and statistical significance was determined by log-rank test (n = 5 mice in each group).
Supplementary information
Supplementary Information
Supplementary Figure 1 and Supplementary Tables 1–8
Source data
Source Data Fig. 1
Unprocessed Western Blots
Source Data Fig. 2
Unprocessed Western Blots
Source Data Fig. 3
Unprocessed Western Blots
Source Data Fig. 4
Unprocessed Western Blots
Source Data Fig. 4
Statistical Source Data
Source Data Fig. 5
Unprocessed Western Blots
Source Data Fig. 5
Statistical Source Data
Source Data Fig. 6
Statistical Source Data
Source Data Extended Data Fig. 1
Unprocessed Western Blots
Source Data Extended Data Fig. 2
Unprocessed Western Blots
Source Data Extended Data Fig. 4
Unprocessed Western Blots
Source Data Extended Data Fig. 5
Unprocessed Western Blots
Source Data Extended Data Fig. 6
Unprocessed Western Blots
Source Data Extended Data Fig. 8
Unprocessed Western Blots
Source Data Extended Data Fig. 8
Statistical Source Data
Source Data Extended Data Fig. 10
Statistical Source Data
Rights and permissions
About this article
Cite this article
Khan, S., Zhang, X., Lv, D. et al. A selective BCL-XL PROTAC degrader achieves safe and potent antitumor activity. Nat Med 25, 1938–1947 (2019). https://doi.org/10.1038/s41591-019-0668-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-019-0668-z
This article is cited by
-
Tumor-targeted PROTAC prodrug nanoplatform enables precise protein degradation and combination cancer therapy
Acta Pharmacologica Sinica (2024)
-
Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL
Nature Communications (2024)
-
BCL-XL inhibitors enhance the apoptotic efficacy of BRAF inhibitors in BRAFV600E colorectal cancer
Cell Death & Disease (2024)
-
PROTAC’ing oncoproteins: targeted protein degradation for cancer therapy
Molecular Cancer (2023)
-
Delivering on the promise of protein degraders
Nature Reviews Drug Discovery (2023)