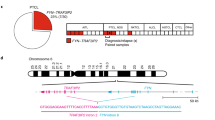
Abstract
Histiocytoses are clonal hematopoietic disorders frequently driven by mutations mapping to the BRAF and MEK1 and MEK2 kinases. Currently, however, the developmental origins of histiocytoses in patients are not well understood, and clinically meaningful therapeutic targets outside of BRAF and MEK are undefined. In this study, we uncovered activating mutations in CSF1R and rearrangements in RET and ALK that conferred dramatic responses to selective inhibition of RET (selpercatinib) and crizotinib, respectively, in patients with histiocytosis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
WES data have been deposited in dbGaP under project accession number phs001864.v1.p1. Source data for Fig. 2 and Extended Data Fig. 10 are presented with this dbGaP paper.
References
Badalian-Very, G. et al. Blood 116, 1919–1923 (2010).
Chakraborty, R. et al. Mutually exclusive recurrent somatic mutations in MAP2K1 and BRAF support a central role for ERK activation in LCH pathogenesis. Blood 124, 3007–3015 (2014).
Diamond, E. L. et al. Diverse and targetable kinase alterations drive histiocytic neoplasms. Cancer Dis. 6, 154–165 (2016).
Hyman, D. M. et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N. Engl. J. Med. 373, 726–736 (2015).
Diamond, E. L. et al. Efficacy of MEK inhibition in patients with histiocytic neoplasms. Nature 567, 521–524 (2019).
Katz, A. M. et al. Langerhans cell histiocytosis in monozygotic twins. J. Am. Acad. Dermatol. 24, 32–37 (1991).
Chantorn, R., Wisuthsarewong, W., Aanpreung, P., Sanpakit, K. & Manonukul, J. Severe congenital systemic juvenile xanthogranuloma in monozygotic twins. Pediatr. Dermatol. 25, 470–473 (2008).
Dai, X. M. et al. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood 99, 111–120 (2002).
Mass, E. et al. A somatic mutation in erythro-myeloid progenitors causes neurodegenerative disease. Nature 549, 389–393 (2017).
Cambiaghi, S., Restano, L. & Caputo, R. Juvenile xanthogranuloma associated with neurofibromatosis 1: 14 patients without evidence of hematologic malignancies. Pediatr. Dermatol. 21, 97–101 (2004).
Elegheert, J. et al. Extracellular complexes of the hematopoietic human and mouse CSF-1 receptor are driven by common assembly principles. Structure 19, 1762–1772 (2011).
Felix, J. et al. Human IL-34 and CSF-1 establish structurally similar extracellular assemblies with their common hematopoietic receptor. Structure 21, 528–539 (2013).
Felix, J. et al. Structure and assembly mechanism of the signaling complex mediated by human CSF-1. Structure 23, 1621–1631 (2015).
Elegheert, J. et al. Allosteric competitive inactivation of hematopoietic CSF-1 signaling by the viral decoy receptor BARF1. Nat. Struct. Mol. Biol. 19, 938–947 (2012).
Walter, M. et al. The 2.7 Å crystal structure of the autoinhibited human c-Fms kinase domain. J. Mol. Biol. 367, 839–847 (2007).
Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J. Mol. Diagn. 17, 251–264 (2015).
Durham, B. H. et al. Genomic analysis of hairy cell leukemia identifies novel recurrent genetic alterations. Blood 130, 1644–1648 (2017).
Benayed, R. et al. High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no mitogenic driver alteration detected by DNA sequencing and low tumor mutation burden. Clin. Cancer Res. 25, 4712–4722 (2019).
Gruber, T. A. et al. An Inv(16)(p13.3q24.3)-encoded CBFA2T3–GLIS2 fusion protein defines an aggressive subtype of pediatric acute megakaryoblastic leukemia. Cancer Cell 22, 683–697 (2012).
Zhang, J. et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481, 157–163 (2012).
Rosenthal, R., McGranahan, N., Herrero, J., Taylor, B. S. & Swanton, C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 17, 31 (2016).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Huang, M. N. et al. MSIseq: software for assessing microsatellite instability from catalogs of somatic mutations. Sci. Rep. 5, 13321 (2015).
Cortes-Ciriano, I., Lee, S., Park, W. Y., Kim, T. M. & Park, P. J. A molecular portrait of microsatellite instability across multiple cancers. Nat. Comm. 8, 15180 (2017).
Alexandrov, L. B. et al. The repertoire of mutational signatures in human cancer. Preprint at bioRxiv https://doi.org/10.1101/322859 (2018).
Walters, D. K. et al. Activating alleles of JAK3 in acute megakaryoblastic leukemia. Cancer Cell 10, 65–75 (2006).
Cammenga, J. et al. Extracellular KIT receptor mutants, commonly found in core binding factor AML, are constitutively active and respond to imatinib mesylate. Blood 106, 3958–3961 (2005).
Maxson, J. E. et al. The colony-stimulating factor 3 receptor T640N mutation is oncogenic, sensitive to JAK inhibition, and mimics T618I. Clin. Cancer Res. 22, 757–764 (2016).
Acknowledgements
We thank the patients and their families for participating in this study, as well as L. Schmitt of UPMC Children’s Hospital of Pittsburgh for histologic technical support. This work was supported by Genentech and grants from the Histiocytosis Association, the Erdheim–Chester Disease Global Alliance, the American Society of Hematology, the Leukemia & Lymphoma Society, the Pershing Square Sohn Foundation, the Functional Genomics Initiative of Memorial Sloan Kettering Cancer Center, the Society of Memorial Sloan Kettering, the Translational and Integrative Medicine Award of Memorial Sloan Kettering, the Geoffrey Beene Center of Memorial Sloan Kettering, the Frame Fund, Nonna’s Garden Foundation, the Flanders Institute for Biotechnology–Belgium and the National Institutes of Health (K08CA218901, UL1TR001857, P30CA008748 and 1R01CA201247).
Author information
Authors and Affiliations
Contributions
B.H.D., E.L.-R., E.L.D. and O.A.-W. designed the study. B.H.D., S.X.L., A.Y., C.E., R.S. and M.K. performed laboratory experiments. S.D.M., E.P. and S.N.S. performed structural studies. B.H.D., E.L.-R., J.P., D.A., V.R., G.A.U., V.S.-M.L., J.M., J.H., J.-F.E. and O.D. collected data and samples. E.L.-R., D.A., M.W., D.M.H., M.E.L., I.D., V.S.-M.L., J.M., J.H., J.-F.E., O.D., A.D. and E.L.D. treated the patients. A.P., D.M., O.C.-B., D.B.S. and M.F.B. performed computational and statistical analyses. G.A.U. evaluated all radiographic studies. B.H.D., J.P., M.Y., K.P.-D., M.E.A., M.L. and J.-F.E. analyzed pathology data. B.H.D., F.G., E.L.D. and O.A.-W. prepared the manuscript with help from all coauthors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare the following competing interests. G.A.U. received personal fees from Sanofi and grants from Sanofi, Novartis and Genentech. M.L. received advisory board compensation from Merck, AstraZeneca, Bristol–Myers Squibb, Takeda and Bayer and research support from Loxo Oncology and Helsinn Healthcare. D.B.S. served as a consultant and received honoraria from Pfizer, Loxo Oncology, Lilly Oncology, Vivideon Therapeutics and Illumina and stock options from Loxo Oncology. M.F.B. received personal fees from Roche and research support from Illumina. D.M.H. received personal fees from Atara Biotherapeutics, Chugai Pharma, Boehringer Ingelheim, AstraZeneca, Pfizer, Bayer, Debiopharm Group and Genentech and grants from the National Cancer Institute, AstraZeneca, Puma Biotechnology and Loxo Oncology. I.D. served as a consultant for Apexigen, Bayer and Celgene and received research support from Bristol–Myers Squibb and Novartis. M.E.L. received personal fees from Loxo Oncology, AstraZeneca, Roche/Genentech and Novartis. A.D. received research funding from Foundation Medicine and personal fees from Ignyta/Genentech/Roche, Loxo/Bayer/Lilly, Takeda/Ariad/Millenium, TP Therapeutics, AstraZeneca, Pfizer, Blueprint Medicines, Helsinn, Beigene, BergenBio, Hengrui Therapeutics, Exelixis, Tyra Biosciences, Verastem and MORE Health. O.A.-W. received grants from H3B Biomedicine and personal fees from H3B Biomedicine, Foundation Medicine, Merck and Jansen unrelated to this manuscript. The remaining authors have nothing to disclose.
Additional information
Peer review information Javier Carmona was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Pedigree and genomic analyses of monozygotic twins with CSF-1R mutant Juvenile Xanthogranuloma (JXG).
(a) Pedigree of twins with JXG. Twin 1 and 2 are monochorionic, diamniotic identical twins born to an otherwise healthy 35-year-old mother. At one year of age, both twins developed small skin lesions on their forehead, which over the course of four months grew in number and diameter extending to the scalp and upper chest. Scalp biopsies were consistent with a diagnosis of JXG. Ophthalmic examination of both twins also identified multiple, bilateral subconjunctival lesions compatible with JXG in twin 1 only. Both twins were monitored in our institution until 2.5 years-of-age with stable, persistent JXG with no visual compromise or neurologic signs or symptoms, and therefore no indication for therapy. Complete blood counts and comprehensive metabolic profiles of both twins have been within normal limits since birth. (b) Mutational profile of the JXG lesion from each twin displaying the fraction of mutations found in each trinucleotide context (performed using deconstructSigs21). (c) Piechart showing the relative contribution of each mutational signature in the JXG lesion from (b) in each twin (based on the 30 mutational signatures detected by Sanger/COSMIC25). Signatures 6 and 15 are associated with defective DNA mismatch repair25. (d) Histogram illustrating the fraction of indels among all mutations identified in the JXG lesions of twins 1 and 2 (red dashed lines) along with the distribution of indel fractions for TCGA samples (colorectal, endometrial, ovarian, stomach) with known microsatellite instability (MSI) status assayed from mononucleotide markers (as described previously23,24; MSI-H: high microsatellite instability; MSS: microsatellite stable). (e) Bar graph depicting proportion of histiocytosis samples that are MSS (90%; n = 28/31) versus MSI-H (10%; n = 3/31) based on WES.
Extended Data Fig. 2 Pie charts of histologic subsets of histiocytoses analyzed with kinase mutations identified in each subset.
(a) Numbers and percentage of patients with each histological subtype of histiocytosis sequenced in this study. Frequency of kinase mutations in the subset of patients with (b) Erdheim-Chester Disease, (c) Langerhans Cell Histiocytosis, (d) Juvenile Xanthogranuloma, (e) Rosai-Dorfman Disease, and (f) Histiocytic Sarcoma.
Extended Data Fig. 3 CSF-1R mutants are expressed on the cell surface and result in phosphorylation of CSF-1R.
(a) Representative anti-CSF-1R and GFP flow cytometry analysis of Ba/F3 cell expressions of human CSF-1R constructs from an MSCV-IRES-GFP construct. Experiments were performed with n = 3 independent biological replicates. (b) Representative histograms of the median fluorescent intensity (MFI) of phospho-CSF-1R Tyrosine 723 intracellular flow cytometry in cells from (a) in cytokine depletion conditions following 60 minutes of stimulation with PBS, human M-CSF (hM-CSF; 100 ng/mL), or hIL-34 (100 ng/mL). Cells expressing empty vector (‘vector’) are shown in the top three rows of each plot) and those expressing wild-type (WT) or mutant CSF-1R constructs are shown in the bottom three rows of each plot. Experiments were performed with n = 3 independent biological experiments. (c) Bar graphs of IC50 values of Ba/F3 cells expressing an empty vector control or CSF-1R mutations in response to Pexidartinib (left) or BLZ945 (right). Mean values of n = 3 biologically independent experiments ± standard deviation is shown. Calculation of p-values performed using ordinary one-way ANOVA; ****p < 0.0001.
Extended Data Fig. 4 Novel kinase mutations and fusions identified in patients with histiocytic neoplasms in this study.
(a) Protein diagrams of novel, somatic point mutations uncovered in kinases in histiocytoses in this study, as well as whether they have been previously described as somatic in cancer (noted by an asterisk, ‘*’) and/or functionally characterized (noted by pound sign, ‘#’).Of the 14 mutations illustrated, the RAF1 K106N5, MEK2 Y134H5, JAK3 V722I26, KIT V530I27, and CSF3R T64028 mutations have been shown to be activating previously. Also, four co-existed with other kinase alterations in the following combinations: CSF3R R583H mutation co-occurred with CSF3R T640I in a JXG patient, KIT R888W mutation co-occurred with MEK1 F53L in a histiocytic sarcoma patient, KIT V530I mutations co-occurred with a MEK1 V93I mutation in a JXG patient. (b) BRAF fusions, (c) NTRK1 fusions, (d) and ALK fusions identified. Hematoxylin and eosin (H&E) and anti-NTRK1 and anti-ALK immunohistochemical staining shown in TPM3-NTRK1-rearranged juvenile xanthogranuloma and KIF5B-ALK-rearranged Erdheim-Chester Disease tumor biopsies, respectively.
Extended Data Fig. 5 Oncoprint of mutations identified in the Erdheim-Chester Disease cohort (n = 100 patients).
Results of whole exome and targeted DNA and RNA sequencing of non-LCH neoplasms. Each patient is represented in one column. Diagnosis (ECD), age category, and sequencing method are in the first 3 rows. Somatic mutations identified are in the lower rows and subdivided based on mutations known to activate kinases, affect the JNK/p38 MAP kinase pathway, or involve a diverse array of co-occurring pathways (shown on the right).
Extended Data Fig. 6 Oncoprint of mutations identified in the Langerhans Cell Histiocytosis cohort (n = 92 patients).
Results of whole exome and targeted DNA and RNA sequencing of LCH neoplasms. Each patient is represented in one column. Diagnosis (LCH), age category, and sequencing method are in the first 3 rows. Somatic mutations identified are in the lower rows and subdivided based on mutations known to activate kinases, affect the JNK/p38 MAP kinase pathway, or involve a diverse array of co-occurring pathways (shown on the right).
Extended Data Fig. 7 Oncoprint of mutations identified in the Juvenile Xanthogranuloma cohort (n = 55 patients).
Results of whole exome and targeted DNA and RNA sequencing of non-LCH neoplasms. Each patient is represented in one column. Diagnosis (JXG), age category, and sequencing method are in the first 3 rows. Somatic mutations identified are in the lower rows and subdivided based on mutations known to activate kinases, affect the JNK/p38 MAP kinase pathway, or involve a diverse array of co-occurring pathways (shown on the right).
Extended Data Fig. 8 Oncoprint of mutations identified in the Rosai Dorfman Disease cohort (n = 17 patients).
Results of whole exome and targeted DNA and RNA sequencing of non-LCH neoplasms. Each patient is represented in one column. Diagnosis (RDD), age category, and sequencing method are in the first 3 rows. Somatic mutations identified are in the lower rows and subdivided based on mutations known to activate kinases or involve a diverse array of co-occurring pathways (shown on the right).
Extended Data Fig. 9 Mutations identified in histiocytic sarcoma (HS) (n = 6 patients).
Results of whole exome and targeted DNA and RNA sequencing of non-LCH neoplasms. Each patient is represented in one column. Diagnosis (HS), age category, and sequencing method are in the first 3 rows. Somatic mutations identified are in the lower rows and subdivided based on mutations known to activate kinases or involve a diverse array of co-occurring pathways (shown on the right).
Extended Data Fig. 10 Characteristics of RET fusion-driven histiocytosis and response to selpercatinib.
(a) Protein diagram of NCOA4-RET fusion identified in a cutaneous xanthogranuoma patient. Photographs of NCOA4-RET JXG skin lesions pre- and 12-weeks post selpercatinib on back (b), scrotum (c), and neck (d). (e) Number of Ba/F3 cells expressing an empty vector or the NCOA4-RET fusion following IL-3 withdrawal (mean of n = 3 independent biological experiments ± standard deviation). Calculation of p-values performed using two-way ANOVA; ****p < 0.0001. Representative western blotting (f) and phospho-protein flow cytometric analysis (g) of phospho-MEK1/2 and phosho-ERK1/2 in the cells from (e). Experiments performed in three independent biological experiments with similar results.
Supplementary information
Supplementary Information
Supplementary Tables 1–6
Source Data Fig. 2
Unprocessed western blots
Source Data Extended Data Fig. 10
Unprocessed western blots
Rights and permissions
About this article
Cite this article
Durham, B.H., Lopez Rodrigo, E., Picarsic, J. et al. Activating mutations in CSF1R and additional receptor tyrosine kinases in histiocytic neoplasms. Nat Med 25, 1839–1842 (2019). https://doi.org/10.1038/s41591-019-0653-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-019-0653-6
This article is cited by
-
Molecular Mutations in Histiocytosis: A Comprehensive Survey of Genetic Alterations
Molecular Biotechnology (2024)
-
A rare case of primary central nervous system histiocytic sarcoma harboring a novel ARHGAP45::BRAF fusion: a case report and literature review
Brain Tumor Pathology (2024)
-
Protein kinases: drug targets for immunological disorders
Nature Reviews Immunology (2023)
-
CSF1R inhibition for histiocytic neoplasm with CBL mutations refractory to MEK1/2 inhibition
Leukemia (2023)
-
OCT2 expression in histiocytoses
Virchows Archiv (2023)