Abstract
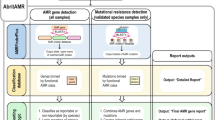
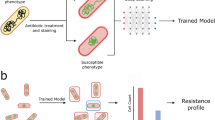
Multidrug resistant organisms are a serious threat to human health1,2. Fast, accurate antibiotic susceptibility testing (AST) is a critical need in addressing escalating antibiotic resistance, since delays in identifying multidrug resistant organisms increase mortality3,4 and use of broad-spectrum antibiotics, further selecting for resistant organisms. Yet current growth-based AST assays, such as broth microdilution5, require several days before informing key clinical decisions. Rapid AST would transform the care of patients with infection while ensuring that our antibiotic arsenal is deployed as efficiently as possible. Growth-based assays are fundamentally constrained in speed by doubling time of the pathogen, and genotypic assays are limited by the ever-growing diversity and complexity of bacterial antibiotic resistance mechanisms. Here we describe a rapid assay for combined genotypic and phenotypic AST through RNA detection, GoPhAST-R, that classifies strains with 94–99% accuracy by coupling machine learning analysis of early antibiotic-induced transcriptional changes with simultaneous detection of key genetic resistance determinants to increase accuracy of resistance detection, facilitate molecular epidemiology and enable early detection of emerging resistance mechanisms. This two-pronged approach provides phenotypic AST 24–36 h faster than standard workflows, with <4 h assay time on a pilot instrument for hybridization-based multiplexed RNA detection implemented directly from positive blood cultures.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All RNA-seq data generated and analyzed for this study, supporting the analyses in Fig. 1 and Extended Data Figs. 1, 2 and 5, have been deposited as aligned bam files in the NCBI Sequencing Read Archive under study ID PRJNA518730. All other datasets generated during the current study, including raw and processed NanoString data, are available from the corresponding author on reasonable request.
Code availability
Custom scripts for transcript selection from RNA-seq data are available at https://github.com/broadinstitute/GeneSelection/. Custom scripts for feature selection and strain classification from NanoString data are available at https://github.com/broadinstitute/DecisionAnalysis/. See the Nature Research Reporting Summary for additional detail.
References
Fauci, A. S. & Morens, D. M. The perpetual challenge of infectious diseases. N. Engl. J. Med. 366, 454–461 (2012).
Antimicrobial Resistance: Global Report on Surveillance 2014 (World Health Organization, 2014).
Kumar, A. et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 34, 1589–1596 (2006).
Kadri, S. S. et al. Difficult-to-treat resistance in gram-negative bacteremia at 173 US hospitals: retrospective cohort analysis of prevalence, predictors, and outcome of resistance to all first-line agents. Clin. Infect Dis. 67, 1803–1814 (2018).
Wiegand, I., Hilpert, K. & Hancock, R. E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175 (2008).
Evans, S. R. et al. Rapid molecular diagnostics, antibiotic treatment decisions, and developing approaches to inform empiric therapy: PRIMERS I and II. Clin. Infect Dis. 62, 181–189 (2016).
Arzanlou, M., Chai, W. C. & Venter, H. Intrinsic, adaptive and acquired antimicrobial resistance in Gram-negative bacteria. Essays Biochem. 61, 49–59 (2017).
Cerqueira, G. C. et al. Multi-institute analysis of carbapenem resistance reveals remarkable diversity, unexplained mechanisms, and limited clonal outbreaks. Proc. Natl Acad. Sci. USA 114, 1135–1140 (2017).
Milheirico, C., de Lencastre, H. & Tomasz, A. Full-genome sequencing identifies in the genetic background several determinants that modulate the resistance phenotype in methicillin-resistant Staphylococcus aureus strains carrying the novel mecC gene. Antimicrob. Agents Chemother. 61, e02500-16 (2017).
Burnham, C. D., Leeds, J., Nordmann, P., O'Grady, J. & Patel, J. Diagnosing antimicrobial resistance. Nat. Rev. Microbiol. 15, 697–703 (2017).
Consortium, C. R. et al. Prediction of susceptibility to first-line tuberculosis drugs by DNA sequencing. N. Engl. J. Med. 379, 1403–1415 (2018).
Jia, B. et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573 (2017).
Bhattacharyya, R. P., Grad, Y. H. & Hung, D. T. in Harrison’s Principles of Internal Medicine (eds J. L. Jameson et al.) Ch. 474 (McGraw-Hill Education, 2018).
Ellington, M. J. et al. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee. Clin. Microbiol. Infect. 23, 2–22 (2017).
Charnot-Katsikas, A. et al. Use of the accelerate pheno system for identification and antimicrobial susceptibility testing of pathogens in positive blood cultures and impact on time to results and workflow. J. Clin. Microbiol. 56, e01166-17 (2018).
Cermak, N. et al. High-throughput measurement of single-cell growth rates using serial microfluidic mass sensor arrays. Nat. Biotechnol. 34, 1052–1059 (2016).
Barczak, A. K. et al. RNA signatures allow rapid identification of pathogens and antibiotic susceptibilities. Proc. Natl Acad. Sci. USA 109, 6217–6222 (2012).
Quach, D. T., Sakoulas, G., Nizet, V., Pogliano, J. & Pogliano, K. Bacterial cytological profiling (BCP) as a rapid and accurate antimicrobial susceptibility testing method for Staphylococcus aureus. EBioMedicine 4, 95–103 (2016).
van Belkum, A., Welker, M., Pincus, D., Charrier, J. P. & Girard, V. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry in clinical microbiology: what are the current issues? Ann. Lab. Med. 37, 475–483 (2017).
Bonomo, R. A. et al. Carbapenemase-producing organisms: a global scourge. Clin. Infect. Dis. 66, 1290–1297 (2018).
Lutgring, J. D. & Limbago, B. M. The problem of carbapenemase-producing-carbapenem-resistant-Enterobacteriaceae detection. J. Clin. Microbiol. 54, 529–534 (2016).
Weisenberg, S. A., Morgan, D. J., Espinal-Witter, R. & Larone, D. H. Clinical outcomes of patients with Klebsiella pneumoniae carbapenemase-producing K. pneumoniae after treatment with imipenem or meropenem. Diagn. Microbiol. Infect. Dis. 64, 233–235 (2009).
Woodworth, K. R. et al. Vital signs: containment of novel multidrug-resistant organisms and resistance mechanisms—United States, 2006–2017. MMWR Morb. Mortal. Wkly Rep. 67, 396–401 (2018).
McMullen, A. R., Yarbrough, M. L., Wallace, M. A., Shupe, A. & Burnham, C. D. Evaluation of genotypic and phenotypic methods to detect carbapenemase production in gram-negative bacilli. Clin. Chem. 63, 723–730 (2017).
Humphries, R. M. CIM City: the game continues for a better carbapenemase test. J. Clin. Microbiol. 57, e00353-19 (2019).
M100: Performance Standards for Antimicrobial Susceptibility Testing 28th edn (Clinical and Laboratory Standards Institute, 2018).
Shishkin, A. A. et al. Simultaneous generation of many RNA-seq libraries in a single reaction. Nat. Methods 12, 323–325 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Geiss, G. K. et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 26, 317–325 (2008).
M07: Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically 11th edn (Clinical and Laboratory Standards Institute, 2018).
Smith, K. P. & Kirby, J. E. The inoculum effect in the era of multidrug resistance: minor differences in inoculum have dramatic effect on MIC determination. Antimicrob. Agents Chemother. 62, e00433-18 (2018).
Adler, A., Ben-Dalak, M., Chmelnitsky, I. & Carmeli, Y. Effect of resistance mechanisms on the inoculum effect of carbapenem in Klebsiella pneumoniae isolates with borderline carbapenem resistance. Antimicrob. Agents Chemother. 59, 5014–5017 (2015).
Nordmann, P., Dortet, L. & Poirel, L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol. Med. 18, 263–272 (2012).
Cubero, M. et al. Carbapenem-resistant and carbapenem-susceptible isogenic isolates of Klebsiella pneumoniae ST101 causing infection in a tertiary hospital. BMC Microbiol. 15, 177 (2015).
Ma, P., Laibinis, H. H., Ernst, C. M. & Hung, D. T. Carbapenem resistance caused by high-level expression of OXA-663 beta-lactamase in an OmpK36-deficient Klebsiella pneumoniae clinical isolate. Antimicrob. Agents Chemother. 62, e01281-18 (2018).
Hou, H. W., Bhattacharyya, R. P., Hung, D. T. & Han, J. Direct detection and drug-resistance profiling of bacteremias using inertial microfluidics. Lab Chip 15, 2297–2307 (2015).
Lomovskaya, O. et al. Vaborbactam: Spectrum of beta-lactamase inhibition and impact of resistance mechanisms on activity in Enterobacteriaceae. Antimicrob. Agents Chemother. 61, e01443-17 (2017).
Marshall, S. et al. Can ceftazidime-avibactam and aztreonam overcome beta-lactam resistance conferred by metallo-beta-lactamases in Enterobacteriaceae? Antimicrob. Agents Chemother. 61, e02243-16 (2017).
Caniaux, I., van Belkum, A., Zambardi, G., Poirel, L. & Gros, M. F. MCR: modern colistin resistance. Eur. J. Clin. Microbiol. Infect. Dis. 36, 415–420 (2017).
Florio, W., Tavanti, A., Barnini, S., Ghelardi, E. & Lupetti, A. Recent advances and ongoing challenges in the diagnosis of microbial infections by MALDI-TOF mass spectrometry. Front. Microbiol. 9, 1097 (2018).
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Gotz, S. et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435 (2008).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, RESEARCH0034 (2002).
Brown, L. D., Cai, T. T. & DasGupta, A. Interval estimation for a binomial proportion. Statist. Sci. 16, 101–133 (2001).
Robnik-Šikonja, M. & Kononenko, I. Theoretical and empirical analysis of ReliefF and RReliefF. Mach. Learn. 53, 23–69 (2003).
Liaw, A. & Wiener, M. Classification and regression by randomForest. R News 2, 18–22 (2002).
Efron, B. & Gong, G. A leisurely look at the bootstrap, the jackknife, and cross-validation. Am. Stat. 37, 36–48 (1983).
Acknowledgements
We thank T. Abeel for valuable early discussions about the approach to machine learning for feature selection and strain classification; E. Lander, B. Birren, C. Nusbaum and C. Russ for input on RNA-seq experiments and overall strategy; and G. Giannoukos and D. Ciulla for valuable contributions to RNA-seq methods development. This publication was supported in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (award nos. 1R01AI117043-05 to D.T.H. and 1K08AI119157-04 to R.P.B., and contract no. HHSN272200900018C). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
R.P.B., J. Livny and D.T.H. conceived of the approach and designed experiments. R.P.B., S.S.S., J. Liu, R.F.R. and M.M.P. designed and executed the RNA-seq experiments. R.P.B., N.B., R.Y., N.S. and J. Livny planned and implemented the RNA-seq analysis, transcript selection and strain classification. R.P.B. analyzed strain phylogenies. R.P.B., P.M., G.C.C., A.P., A.M.E., J. Livny and D.T.H. planned and implemented the strategy for carbapenemase and ESBL gene detection. R.P.B., S.S.S., J. Liu and L.L.H. executed the NanoString experiments. R.P.B., J.S., V.M.P., L.L.H. and L.A.C. designed and executed clinical sample collection and processing for real and simulated blood cultures. E.N., K.A.M. and J.T. assisted with experimental design and sample acquisition for genotypic and phenotypic analysis of carbapenem-resistant clinical isolates. L.W., R.K., R.B. and J.B. designed and implemented the Hyb & Seq experiments. R.P.B. and D.T.H. primarily drafted the manuscript, with extensive input from J. Livny. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
R.P.B., P.M., J. Livny and D.T.H. are co-inventors on subject matter in US provisional application No. 62/723,417 filed by the Broad Institute directed to RNA signatures for AST, as described in this manuscript. L.W., R.B., R.K. and J.B. are employees at NanoString, Inc., the company that manufactures the RNA detection platforms used in this manuscript. NanoString, Inc. has licensed the intellectual property for RNA-based AST from the Broad Institute. V.M.P. received research funds from SeLux Diagnostics, Inc. for work on an unrelated project.
Additional information
Peer review information Alison Farrell was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Differential gene expression upon antibiotic exposure distinguishes susceptible and resistant strains.
(a) RNA-Seq data from two susceptible (left panels) or two resistant (right panels) clinical isolates of E. coli or A. baumannii treated with meropenem (60 min), ciprofloxacin (30 min), or gentamicin (60 min) at CLSI breakpoint concentrations are presented as MA plots. Statistical significance was determined by a two-sided Wald test with the Benjamini-Hochberg correction for multiple hypothesis testing, using the DESeq2 package28. (b) Heatmaps of normalized, log-transformed fold-induction of top 10 antibiotic-responsive transcripts from 24 clinical isolates of E. coli or A. baumannii treated at CLSI breakpoint concentrations with meropenem, ciprofloxacin, or gentamicin. Gene identifiers are listed at right, along with gene names if available. CLSI classifications of each strain based on broth microdilution are shown below. * = strains with large inoculum effects in meropenem MIC; x = strains discordant by more than one dilution. (c) GoPhAST-R predictions of probability of resistance from a random forest model trained on NanoString data from the derivation cohort and tested on the validation cohort (y-axis) are compared with standard CLSI classification based on broth microdilution MIC (x-axis) for E. coli (top) or A. baumannii isolates treated with meropenem, ciprofloxacin, and gentamicin. Horizontal dashed lines indicate 50% probability of resistance. Vertical dashed lines indicate the CLSI breakpoint between susceptible and not susceptible (that is intermediate/resistant). Numbers in each quadrant indicate concordant and discordant classifications between GoPhAST-R and broth microdilution. Carbapenemase (square outline) and select ESBL (diamond outline) gene content as detected by GoPhAST-R are also displayed on the meropenem plot.
Extended Data Fig. 2 Timecourse of RNA-Seq data upon antibiotic exposure reveals differential gene expression between susceptible and resistant clinical isolates.
Susceptible (left panels) or resistant (right panels) clinical isolates of K. pneumoniae, E. coli, or A. baumannii treated with meropenem, ciprofloxacin, or gentamicin at CLSI breakpoint concentrations for the indicated times. Data are presented as MA plots, Statistical significance was determined by a two-sided Wald test with the Benjamini-Hochberg correction for multiple hypothesis testing, using the DESeq2 package28.
Extended Data Fig. 3 Phylogenetic trees highlight the diversity of strains used in this study.
Phylogenetic trees of all sequenced isolates deposited in NCBI for (a) K. pneumoniae, (b) E. coli, (c) A. baumannii, and (d) P. aeruginosa, with all sequenced isolates used in this study indicated by colored arrowheads around the periphery. See Supplementary Methods for details.
Extended Data Fig. 4 NanoString data from dozens of antibiotic-responsive genes distinguish susceptible from resistant isolates.
Heatmaps of normalized, log-transformed fold-induction of antibiotic-responsive transcripts from clinical isolates of K. pneumoniae (24, 18, and 26 independent clinical isolates for the three antibiotics, respectively), E. coli (24 independent clinical isolates for each antibiotic), or A. baumannii (24 clinical isolates for each antibiotic) treated at CLSI breakpoint concentrations with meropenem, ciprofloxacin, or gentamicin. CLSI classifications are shown below. All antibiotic-responsive transcripts chosen as described from RNA-Seq data are shown here; the subset of these chosen by reliefF as the 10 most discriminating transcripts are shown in Fig. 1b or Supplemental Fig. 1b. * = strains with large inoculum effects in meropenem MIC; + = one-dilution errors; x = strains discordant by more than one dilution.
Extended Data Fig. 5 One-dimensional projection of NanoString data distinguishes susceptible from resistant isolates and reflects MIC.
(a) Phase 1 NanoString data from Supplemental Fig. 2 (that is, normalized, log-transformed fold-induction for each responsive transcript), analyzed as described to generate squared projected distance (SPD) metrics (y-axes) for each strain (see Supplementary Methods), are binned by CLSI classifications (x-axes), for clinical isolates of K. pneumoniae (24, 18, and 26 independent clinical isolates for the three antibiotics, respectively), E. coli (24 independent clinical isolates for each antibiotic), or A. baumannii (24 clinical isolates for each antibiotic) treated at CLSI breakpoint concentrations with meropenem, ciprofloxacin, or gentamicin (the same isolates shown in Fig. 1b, c and Supplemental Fig. 1b, c). By definition, an SPD of 0 indicates a transcriptional response to antibiotic equivalent to that of an average susceptible strain, while an SPD of 1 indicates a response equivalent to that of an average resistant strain. See Supplementary Methods for details. Data are summarized as box-and-whisker plots, where boxes extend from the 25th to 75th percentile for each category, with a line at the median, and whiskers extend from the minimum to the maximum. Note that for A. baumannii and meropenem, the clustering of the majority of susceptible strains by this simple metric (aside from one outlier which is misclassified as resistant by GoPhAST-R) underscores the true differences in transcription between susceptible and resistant isolates, despite the more subtle-appearing differences in heatmaps for this combination (Supplemental Fig. 1b), which is largely caused by one strain with an exaggerated transcriptional response (seen here as the strain with a markedly negative SPD) that affects scaling of the heatmap. (b) The same SPD data (y-axes) plotted against broth microdilution MICs (x-axes) reveal that the magnitude of the transcriptional response to antibiotic exposure correlates with MIC. In both (a) and (b), strains with a large inoculum effect upon meropenem treatment are displayed in red and enlarged. Vertical dashed line indicates the CLSI breakpoint between susceptible and not susceptible (that is, intermediate or resistant).
Extended Data Fig. 6 RNA-Seq and NanoString data reveal differential gene expression that distinguishes susceptible from resistant clinical isolates for S. aureus + levofloxacin and P. aeruginosa + ciprofloxacin.
(a) RNA-Seq data from two susceptible or two resistant clinical isolates of each species treated with the indicated fluoroquinolone at 1 mg/L for 60 minutes are presented as MA plots. Statistical significance was determined by a two-sided Wald test with the Benjamini-Hochberg correction for multiple hypothesis testing, using the DESeq2 package28. (b) Heatmaps of normalized, log-transformed fold-induction of antibiotic-responsive transcripts from 24 independent clinical isolates of each species treated with the indicated fluoroquinolone at 1 mg/L for 60 minutes. For each species, NanoString data from all candidate transcripts are shown at left, and top the 10 transcripts selected from Phase 1 testing are shown at right. (c) GoPhAST-R predictions of probability of resistance from a random forest model trained on Phase 1 NanoString data from the derivation cohort and tested on the validation cohort (y-axis) compared with standard CLSI classification based on broth microdilution MIC (x-axis). Horizontal dashed lines indicate 50% probability of resistance. Vertical dashed lines indicate the CLSI breakpoint between susceptible and not susceptible (that is intermediate/resistant). Numbers in each quadrant indicate concordant and discordant classifications between GoPhAST-R and broth microdilution.
Extended Data Fig. 7 Schematic of data analysis scheme, including ‘two-phase’ machine learning approach to feature selection and strain classification.
Schematic representation of major data analysis steps in identifying antibiotic-responsive transcriptional signatures from RNA-Seq data, validating and optimizing these signatures using NanoString in two phases, and using these signatures to classify strains of unknown MIC, also in two phases. First, candidate antibiotic-responsive and control transcripts were chosen from RNA-Seq data using custom scripts built around the DESeq2 package47, and conserved regions of these transcripts were identified for targeting in a hybridization assay. In phase 1 (implemented for all pathogen-antibiotic pairs), these candidate transcripts were quantitated on the NanoString assay platform, and the resulting data were partitioned by strain into training and testing cohorts. Ten transcripts that best distinguish susceptible from resistant strains within the training cohort were then selected (step 1 A) using the reliefF feature selection algorithm (implemented via the CORElearn package), then used to train an ensemble classifier (step 1B) on the same training cohort using a random forest algorithm (implemented via the caret package). This trained classifier was then used to predict susceptibilities of strains in the testing cohort (step 1 C), and accuracy was assessed by comparing with broth microdilution results (Supplementary Table 4). In phase 2 (implemented for K. pneumoniae + meropenem and ciprofloxacin), the same process was repeated, but the phase 1 training and testing cohorts were combined into a single, larger training cohort for feature selection (step 2 A) and classifier training (step 2B), and a new set of strains were obtained as a testing cohort. The 10 genes selected from the phase 2 training cohort were measured from this phase 2 testing cohort, and the trained classifier was used for AST on these new strains (step 2 C), with accuracy again assessed by comparison with broth microdilution (Supplementary Table 4). See Supplementary Methods for detailed descriptions of each of these analysis steps.
Extended Data Fig. 8 GoPhAST-R accurately classifies K. pneumoniae isolates tested in phase 2.
(a) Heatmaps of normalized, log-transformed fold-induction of top 10 antibiotic-responsive transcripts from K. pneumoniae treated at CLSI breakpoint concentrations with meropenem (31 independent clinical isolates) or ciprofloxacin (25 independent clinical isolates). CLSI classifications are shown below. * = strain with large inoculum effects in meropenem MIC; + = one-dilution error; x = strain discordant by more than one dilution. Note that the 10 responsive transcripts shown are the only 10 tested for this second phase of GoPhAST-R implementation. (b) GoPhAST-R predictions of probability of resistance from a random forest model trained on all Phase 1 NanoString data the independent Phase 2 cohort (y-axis) compared with standard CLSI classification based on broth microdilution MIC (x-axis). Horizontal dashed lines indicate 50% probability of resistance. Vertical dashed lines indicate the CLSI breakpoint between susceptible and not susceptible (that is intermediate/resistant). Numbers in each quadrant indicate concordant and discordant classifications between GoPhAST-R and broth microdilution. * = strain with large inoculum effects in meropenem MIC.
Extended Data Fig. 9 GoPhAST-R accurately classifies AST and detects key resistance elements directly from simulated positive blood culture bottles in < 4 h.
(a) Heatmaps of normalized, log-transformed fold-induction NanoString data from the top 10 antibiotic-responsive transcripts directly from 12 simulated positive blood culture bottles for each indicated pathogen-antibiotic combination reveal antibiotic-responsive transcription in susceptible but not resistant isolates. For meropenem, results of carbapenemase / ESBL gene detection are also displayed as a normalized, background-subtracted, log-transformed heatmap above. CLSI classifications of isolates, which were blinded until analysis was complete, are displayed below each heatmap. (b) Probability of resistance from a random forest model trained by leave-one-out cross-validation on NanoString data from (a) (y-axis) compared with standard CLSI classification based on broth microdilution MIC (x-axis) for each isolate. Horizontal dashed lines indicate 50% chance of resistance based on random forest model. Vertical dashed lines indicate CLSI breakpoint between susceptible and resistant. Carbapenemase (square outline) and select ESBL (diamond outline) gene content as detected by GoPhAST-R are also displayed on meropenem plots. See Supplementary Methods for details of spike-in protocol.
Supplementary information
Rights and permissions
About this article
Cite this article
Bhattacharyya, R.P., Bandyopadhyay, N., Ma, P. et al. Simultaneous detection of genotype and phenotype enables rapid and accurate antibiotic susceptibility determination. Nat Med 25, 1858–1864 (2019). https://doi.org/10.1038/s41591-019-0650-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-019-0650-9
This article is cited by
-
Antimicrobial resistance crisis: could artificial intelligence be the solution?
Military Medical Research (2024)
-
RNA-based antibiotic susceptibility testing of tmexCD-toprJ-mediated tigecycline resistance in Klebsiella pneumoniae
One Health Advances (2023)
-
TRS: a method for determining transcript termini from RNAtag-seq sequencing data
Nature Communications (2023)
-
The membrane-active polyaminoisoprenyl compound NV716 re-sensitizes Pseudomonas aeruginosa to antibiotics and reduces bacterial virulence
Communications Biology (2022)
-
A genome-wide atlas of antibiotic susceptibility targets and pathways to tolerance
Nature Communications (2022)