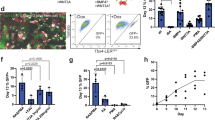
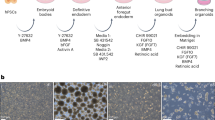
Abstract
Millions of people worldwide with incurable end-stage lung disease die because of inadequate treatment options and limited availability of donor organs for lung transplantation1. Current bioengineering strategies to regenerate the lung have not been able to replicate its extraordinary cellular diversity and complex three-dimensional arrangement, which are indispensable for life-sustaining gas exchange2,3. Here we report the successful generation of functional lungs in mice through a conditional blastocyst complementation (CBC) approach that vacates a specific niche in chimeric hosts and allows for initiation of organogenesis by donor mouse pluripotent stem cells (PSCs). We show that wild-type donor PSCs rescued lung formation in genetically defective recipient mouse embryos unable to specify (due to Ctnnb1cnull mutation) or expand (due to Fgfr2cnull mutation) early respiratory endodermal progenitors. Rescued neonates survived into adulthood and had lungs functionally indistinguishable from those of wild-type littermates. Efficient chimera formation and lung complementation required newly developed culture conditions that maintained the developmental potential of the donor PSCs and were associated with global DNA hypomethylation and increased H4 histone acetylation. These results pave the way for the development of new strategies for generating lungs in large animals to enable modeling of human lung disease as well as cell-based therapeutic interventions4,5,6.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Murphy, S. L., Xu, J., Kochanek, K. D. & Arias, E. Mortality in the United States, 2017. NCHS Data Brief no. 328 (National Center for Health Statistics, 2018).
Petersen, T. H. et al. Tissue-engineered lungs for in vivo implantation. Science 329, 538–541 (2010).
Kotton, D. N. & Morrisey, E. E. Lung regeneration: mechanisms, applications and emerging stem cell populations. Nat. Med. 20, 822–832 (2014).
Matsunari, H. et al. Blastocyst complementation generates exogenic pancreas in vivo in apancreatic cloned pigs. Proc. Natl Acad. Sci. USA 110, 4557–4562 (2013).
Wu, J. et al. Interspecies chimerism with mammalian pluripotent stem cells. Cell 168, 473–486.e15 (2017).
Suchy, F., Yamaguchi, T. & Nakauchi, H. iPSC-derived organs in vivo: challenges and promise. Cell Stem Cell 22, 21–24 (2018).
Valapour, M. et al. OPTN/SRTR 2017 annual data report: lung. Am. J. Transplant. 19, 404–484 (2019).
Chen, Y.-W. et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat. Cell Biol. 19, 542–549 (2017).
Dye, B. R. et al. A bioengineered niche promotes in vivo engraftment and maturation of pluripotent stem cell derived human lung organoids. eLife 5, 1–18 (2016).
Ott, H. C. et al. Regeneration and orthotopic transplantation of a bioartificial lung. Nat. Med. 16, 927–933 (2010).
Rosen, C. et al. Preconditioning allows engraftment of mouse and human embryonic lung cells, enabling lung repair in mice. Nat Med 21, 869–879 (2015).
Stone, K. C., Mercer, R. R., Gehr, P., Stockstill, B. & Crapo, J. D. Allometric relationships of cell numbers and size in the mammalian lung. Am. J. Respir. Cell Mol. BioI. 6, 235–243 (1992).
Crapo, J. D., Barry, B. E., Gehr, P., Bachofen, M. & Weibel, E. R. Cell number and cell characteristics of the normal human lung. Am. Rev. Respir. Dis. 126, 332–337 (1982).
Chen, J., Lansford, R., Stewart, V., Young, F. & Alt, F. W. RAG-2-deficient blastocyst complementation: an assay of gene function in lymphocyte development. Proc. Natl Acad. Sci. USA 90, 4528–4532 (1993).
Kobayashi, T. et al. Generation of rat pancreas in mouse by interspecific blastocyst injection of pluripotent stem cells. Cell 142, 787–799 (2010).
Yamaguchi, T. et al. Interspecies organogenesis generates autologous functional islets. Nature 542, 191–196 (2017).
Usui, J. et al. Generation of kidney from pluripotent stem cells via blastocyst complementation. Am. J. Pathol. 180, 2417–2426 (2012).
Freedman, B. S. Hopes and difficulties for blastocyst complementation. Nephron 139, 42–47 (2018).
Harris-Johnson, K. S., Domyan, E. T., Vezina, C. M. & Sun, X. Beta-catenin promotes respiratory progenitor identity in mouse foregut. Proc. Natl Acad. Sci. USA 106, 16287–16292 (2009).
Goss, A. M. et al. Wnt2/2b and β-catenin signaling are necessary and sufficient to specify lung progenitors in the foregut. Dev. Cell 17, 290–298 (2009).
Sekine, K. et al. Fgf10 is essential for limb and lung formation. Nat. Genet. 21, 138–141 (1999).
De Moerlooze, L. et al. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development 127, 483–492 (2000).
Xu, X. et al. Fibroblast growth factor receptor 2 (FGFR2)-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development 125, 753–765 (1998).
Harris, K. S., Zhang, Z., McManus, M. T., Harfe, B. D. & Sun, X. Dicer function is essential for lung epithelium morphogenesis. Proc. Natl Acad. Sci. USA 103, 2208–2213 (2006).
Yu, K. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development 130, 3063–3074 (2003).
Longmire, T. A. et al. Efficient derivation of purified lung and thyroid progenitors from embryonic stem cells. Cell Stem Cell 10, 398–411 (2012).
Ying, Q. L. et al. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008).
Kretsovali, A., Hadjimichael, C. & Charmpilas, N. Histone deacetylase inhibitors in cell pluripotency, differentiation, and reprogramming. Stem Cells Int. 2012, 184154 (2012).
Stadtfeld, M. et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature 465, 175–181 (2010).
Choi, J. et al. Prolonged Mek1/2 suppression impairs the developmental potential of embryonic stem cells. Nature 548, 219–223 (2017).
Yagi, M. et al. Derivation of ground-state female ES cells maintaining gamete-derived DNA methylation. Nature 548, 224–227 (2017).
Furusawa, T., Ohkoshi, K., Honda, C., Takahashi, S. & Tokunaga, T. Embryonic stem cells expressing both platelet endothelial cell adhesion molecule-1 and stage-specific embryonic antigen-1 differentiate predominantly into epiblast cells in a chimeric embryo1. Biol. Reprod. 70, 1452–1457 (2004).
Leitch, H. G. et al. Naive pluripotency is associated with global DNA hypomethylation. Nat. Struct. Mol. Biol. 20, 311–316 (2013).
Lyko, F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 19, 81–92 (2017).
Brault, V. et al. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128, 1253–1264 (2001).
Ke, M. T. et al. Super-resolution mapping of neuronal circuitry with an index-optimized clearing agent. Cell Rep. 14, 2718–2732 (2016).
Mori, M. et al. Cytoplasmic E2f4 forms organizing centres for initiation of centriole amplification during multiciliogenesis. Nat. Commun. 8, 15857 (2017).
Huang, S. X. L. et al. The in vitro generation of lung and airway progenitor cells from human pluripotent stem cells. Nat. Protoc. 10, 413–425 (2015).
Singer, X. B. D. et al. Flow-cytometric method for simultaneous analysis of mouse lung epithelial, endothelial, and hematopoietic lineage cells. Am. J. Physiol. Lung Cell Mol. Physiol. 310, L796–L801 (2016).
Chapman, H. A. et al. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature 517, 621–625 (2015).
Golde, W. T., Gollobin, P. & Rodriguez, L. L. A rapid, simple, and humane method for submandibular bleeding of mice using a lancet. Lab Anim. (NY) 34, 39–43 (2005).
Mori, M. et al. Notch3-Jagged signaling controls the pool of undifferentiated airway progenitors. Development 142, 258–267 (2015).
Lu, C. et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 352, 844–849 (2016).
Mikami, M. et al. Impaired relaxation of airway smooth muscle in mice lacking the actin-binding protein gelsolin. Am. J. Respir. Cell Mol. Biol. 56, 628–636 (2017).
Acknowledgements
We thank J. Qian, J. Huang, A. Kuforiji and M. Jiang for technical assistance, and D. Kotton and L. Oikonomou (CReM, Boston University) for invaluable reagents. We also thank the helpful scientific input from members of the Cardoso’s lab and the Columbia Center for Human Development (J. Lu, H. Snoeck, S. Huang, J. Que, F. Constantini, J. Bhattacharya and M. Bacchetta) as well as from M. Morimoto (RIKEN, Japan), T. Matozaki (Kobe University), H. Masaki, S. Hamanaka (University of Tokyo) and T. Nishimura (Stanford University). We also thank K. Kennedy for help in editing the manuscript. We would like to acknowledge the support from the CCTI flow cytometry core (LSRII: NIH S10RR027050) and Columbia Stem Cell Initiative (CSCI) Flow Cytometry core (FACS Area). This work was funded by the Department of Defense, PR161857 and NIH-NHLBI 1 R01 HL148223-01 to M.M., CIRM Research Leadership Award, LA1_C12-06917 to H.N., NIH-NHLBI R35-HL135834-01 to W.V.C., Giannandrea Family Dale F. Frey Breakthrough Scientist of the Damon Runyon Foundation (DFS-28-18) and a Pew-Stewart Scholar for Cancer Research to C.L.
Author information
Authors and Affiliations
Contributions
M.M. designed and conducted all experiments; J.A.D. and C.W.E. performed pulmonary function assessment; C.-S.L. and Y.T. supported and performed microinjection and embryo transfer; M.M., P.R. and M.O. maintained mutant mice for the injection; K.F. supported imaging of whole-mount staining; K.F., Y.H., M.O. and M.K. supported and performed hematopoietic cell-colony-formation assay; X.X. and C.L. performed epigenetic experiments; M.M., C.L. and W.V.C wrote the paper; H.N. gave crucial insights on the experiments and the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Michael Basson was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 PSCNkx2-1-GFP rescues lung agenesis in Fgfr2-deficient mutants, but lungs are immature.
a, Schematic of experimental procedure (left) and representative macroscopic view of the lungs and heart (ht) from newborn (P0) mice Fgfr2cnull complemented by donor 2i/LIF-cultured PSCNkx2-1-GFP. Right panel depicts GFP signals in the lungs not present in the heart (outlined). b, Representative GFP expression in lung section of P0 Fgfr2cnull + PSCNkx2-1-GFP: strong signals throughout all epithelial tubes, less prominent in the mesenchyme and its derivatives (Ex. large vessels, center). Asterisk (*) marks distal epithelial tubules unable to form distal saccules resulting in immature non-functional lungs. Immunofluorescence of Nkx2-1 and quantitative analysis confirming extensive double-labeling with GFP (single channels shown in small panels). Graph represents mean ± s.e.m. of % GFP+ lung epithelial cells in five random fields per sample (n = 2 animals). c–f, Representative immunofluorescence and confocal images of lungs double-labeled with GFP and markers of alveolar type 1 (Pdpn) and type 2 (Sftpc) or airway multiciliated (acetylated α-tub) or secretory (Scgb1a1) cells (n = 3–4 per group). Boxed area (f) enlarged in the right panels. b–f, images also displayed as single channels; DAPI in grey. Scale bars: a, b, c–e, f: 1 mm, 20 μm, 10 μm, and 20 μm, respectively.
Extended Data Fig. 2 Effect of PSC cell culture conditions in pluripotency markers.
a, Representative morphology of PSCCAG–GFP colonies and immunofluorescence image of Ssea1 and Pecam expression under the culture conditions listed. b, Representative flow cytometry pseudocolor images (top) and respective histograms (bottom) showing the increased yield of the Ssea1highPecam+ PSCCAG–GFP population and high Ssea1 MFI in cultures treated with VPA/LIF and a2i/VPA/LIF. c, FACS analysis of PSCNkx2-1-GFP cultured on the conditions indicated. Graph shows differences in the yield of Ssea1+Pecam+ (%). Note the effect of the addition of VPA to LIF or a2i/LIF treatment. Bars are mean ± standard error of n = 3 independent experiments in each culture condition. Data were analyzed by one-way ANOVA; differences were significant at **P < 0.01. Scale bars: a = 10 μm.
Extended Data Fig. 3 Complemented distal lung of VPA/LIF-treated PSCCAG–GFP Fgfr2 mutants undergo sacculation.
a Representative immunofluorescence confocal imaging depicting the expression of GFP and markers for alveolar type 2 (Sftpc) in the walls of distal saccules in lung sections of P0 VPA/LIF-treated PSCCAG–GFP Fgfr2cnull mice (left panel, WT P0 littermate). b, the 3D-SIM image of PSCCAG-GFP Fgfr2cnull mice showing at high resolution the staining of alveolar type I surface double-labeled with GFP. Scale bars: a and b,10 μm.
Extended Data Fig. 4 Airway epithelial complementation in Fgfr2cnull PSCCAG–GFPnewborn chimeric pups.
a,b, Immunofluorescence and confocal imaging depicting GFP double-labeling (arrows) with cell differentiation markers Scgb1a1 (secretory), β-tubulin4 (multiciliated) and Cgrp (neuroendocrine) in lung sections of P0 mice from VPA/LIF-treated PSCCAG–GFP + Fgfr2cnull and WT chimeric littermates; small panels in b depict single channels). c, Percentage of GFP+ cells in the lung and tracheal epithelium as determined by quantitative analysis of GFP signals in sections of newborn P0 PSCCAG–GFP Fgfr2cnull animals. Graph, Mean ± s.e.m. of measurements in 5 random fields per section per sample. Student’s t-test; **P < 0.01. Right panel, Representative GFP and Sma immunostaining in a histological section of P0 complemented mutant depicting epithelial signals (arrows) consistently strong in intrapulmonary airways (bracket), but variable or low (asterisks) in the extrapulmonary airway (dashed box) and trachea (Tr). Boxed area enlarged on the right. Sma (alpha-smooth muscle actin) labeling airway smooth muscle. Scale bars: a, b, c: 10 μm, 10 μm, and 20 μm, respectively.
Extended Data Fig. 5 Chimerism in the lung alveolar vascular compartment (a–c) and extrapulmonary organs of animals complemented with VPA/LIF-treated PSCCAG–GFP (d–f).
a, Immunofluorescence and confocal imaging of newborn (P0) WT + PSCCAG–GFP, Fgfr2hetero + PSCCAG–GFP and Fgfr2cnull + PSCCAG–GFP chimeric lungs. Representative image of blood vessels showing GFP-Isolectin B4 double-labeled endothelial cells (arrows). b, Percentage of GFP labeling in endothelial (top) and alveolar type I (bottom) cells in P0 WT + PSCCAG–GFP and Fgfr2cnull + PSCCAG–GFP chimeric lungs as determined by morphometric analysis of sections immunostained with Pecam (top) or Hopx and Pdpn (bottom). Graphs: mean ± s.e.m. of measurements in ten non-overlapping random fields per group (see also Supplementary Fig. 2 and Methods). c, GFP-Sma double-labeling of smooth muscle cells (arrow) in blood vessels (bv) and airways (aw) by immunofluorescence of P0 lungs from PSCCAG–GFP complemented Fgfr2cnull mice. d, Proportion of GFP+ cells in the liver from E15.5 and P0 chimeric mice isolated by FACS. Graph represents mean ± s.e.m. of PSCCAG–GFP-complemented WT, Fgfr2hetero or Fgfr2cnull animals (n = 13, 6, 3 animals, respectively). e, Representative images of GFP expression in liver (left) and kidney (right) from P0 Fgfr2cnull + PSCCAG–GFP (whole-mount and histological sections) and quantitative analysis (bottom) of GFP labeling; graph: mean ± s.e.m. of % GFP+ cells per field in 5 random fields per group. f, Representative images of GFP expression in the intestine from WT or Fgfr2cnull injected with PSCCAG–GFP (histological sections) and graph showing % GFP + (mean ± s.e.m., 5 random fields per 5 per group). Statistical analysis (b,d–f): Student’s t-test; **P < 0.01, NS: statistically non-significant. Scale bars: a, b = 10 μm, 5 μm, respectively; e: top panel: 20 µm, bottom panel: 1 mm; f: 10 μm.
Extended Data Fig. 6 Low chimerism in VPA/LIF-treated PSCsCAG–GFP Fgfr2cnull embryos and defective lung organogenesis.
GFP expression in whole-mount E15.5 embryos and tissues. a, Variable chimerism in E15.5 WT + PSCsCAG–GFP embryos with different levels of GFP signals in the skin, lung (lu) and heart (ht: outlined). b, Unilateral rescue of the lung (boxed) in E15.5 Fgfr2cnull embryos complemented with VPA/LIF PSCsCAG–GFP; note low chimerism in the esophagus (es) and heart. Scale bars: 1 mm.
Extended Data Fig. 7 Comparative effects of a2i/VPA/LIF with other culture conditions in PSC pluripotency and chimerism.
a, Immunofluorescence and quantitative analysis of Ssea1 expression in PSCCAG-–GFP colonies in the various media tested. Top panel, representative images of Ssea1high, Ssea1dim and Ssea1negative colonies. Bottom panel, graphs showing the percent of Ssea1-expressing colonies high, dim or negative as above (left) and percent of Ssea1high colonies (right) in each culture condition. Data are mean ± s.e.m of the number of colonies per field in five non-overlapping random fields per condition. b, IF of Oct4 and SSEA1in PSCCAG–GFP cultured in the conditions indicated. Left: representative image of Oct4 staining. Right: Graph showing Oct4 expression levels in SSEA1high (dots) or Ssea1 dim/negative pooled (empty dots) PSC colonies cultured as indicated. Mean fluorescent intensity (MFI) of Oct4 as assessed by imaging of 35 random colonies per culture condition (ImageJ). Relative MFI per colony shown as the Oct4 MFI of each colony normalized by the average fluorescent intensity of the PSCs cultured in LIF condition. Graphs in a and b depicting the VPA’s ability to enrich for Ssea1high PSCs and the a2i/VPA/LIF effect in enhancing Oct4 expression but no significant difference in Oct4 levels between Ssea1high and Ssea1dim a2i/VPA/LIF-treated PSCs. c, Percentage of chimera formation as determined by analysis of skin/coat color in pups at P0 from blastocysts injected with PSCs cultured in each condition and transferred to foster mothers. Graph represents mean ± s.e.m. of the percentage of chimeric pups generated from the PSCs indicated (number of experiments represented by each point in the graph, see Methods). d) Whole-mount brightfield and GFP images of E15.5 embryos showing the high frequency of GFP-expressing chimera formation from WT hosts injected with a2i/VPA/LIF-treated PSCCAG–GFP (see also Table 1). Statistical analysis: one-way ANOVA (a,b) and Student’s t-test (c); differences considered statistically significant if *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, NS: non-significant. Scale bars: a, b, d = 10 μm, 20 μm, 1 mm respectively.
Extended Data Fig. 8 FACS-based assessment of cell type-specific chimerism in lung and liver from a2i/VPA/LIF -treated Ssea1high PSCCAG-GFP WT, Fgfr2hetero, or Fgfr2cnull hosts.
a, Schematics of the CBC approach (Ssea1high a2i/VPA/LIF-treated PSC donor cells injected into blastocysts hosts, chimeric pups identified at birth), tissue isolation/dissociation and gating strategy for flow cytometry (FACS) analysis of different cell types in the lung. Lung cells (FSC/SCC panels) and singlets (FSC-H/FSC-A) were selected. Within the lung singlets, live cells were gated as an unstained negative control (NC) and hematopoietic (CD45+ versus CD45–). In the CD45– gate, we separated epithelial (Epcam+), endothelial (Pecam+) and other non-endothelial lung mesenchymal cells (Pecam/Epcam double negative). The percentage of GFP+ cells was calculated based on the unstained WT negative control gate. b, FACS analysis of complemented lungs (left panels) and liver (right panel) from Ssea1high a2i/VPA/LIF -treated PSCCAG–GFP + WT (n = 5) Fgfr2hetero (n = 7) or Fgfr2cnull (n = 2) animals showing the percentage of GFP labeling in lung (epithelial, endothelial or non-endothelial lung mesenchymal) cells and liver. Graphs (mean ± s.e.m) depicting GFP labeling in nearly all lung epithelial cells from Fgfr2cnull compared to heterozygous and WT, inconsistent variable labeling in lung endothelial/mesenchymal cells and low proportion of GFP+ cells in liver regardless of the genotype.
Extended Data Fig. 9 Effect of culture conditions in global DNA methylation, DNA methyltransferase gene expression and the patterns of histone modifications by PSCCAG–GFP.
a, Analysis of 5-mC levels of Line-1 repeats in cell homogenates from PSCCAG–GFP cultures in the conditions indicated (n = 3 independent experiments); graph represents mean ± s.e.m. showing significantly lower levels of relative DNA methylation in all conditions compared to LIF. b, Relative levels expression of Dnmt3b and Dnmt3a in PSCCAG–GFP cultures by qPCR analysis; data were normalized by the LIF averaged values and represented as mean ± s.e.m (n = 3 independent experiments). c, Western blot of histone methyltransferases and acetylase from PSCs homogenates (n = 3 per condition); expression levels quantitated by densitometry analyses and normalized by LIF values. d, Immunofluorescence of histone H4ac in the PSC-treated cultures indicated (top: representative images). Mean fluorescent intensity (MFI) per colony as assessed by imaging of 20 random colonies per condition, 5 random fields (ImageJ). Graph (bottom) depicting mean ± s.e.m of MFI confirm the consistently high levels of H4-pan-acetyl expression in a2i/VPA/LIF also seen in Western blots. Statistical analyses: one-way ANOVA (a,b) and unpaired Student’s t-test (d), significance at *P < 0.05, **P < 0.01, ****P < 0.0001, NS: non-significant.
Extended Data Fig. 10 Normal growth, differentiation and function of adult lungs from complemented a2i/VPA/LIF-treated PSCCAG–GFP mature animals (see also Figs. 2 and 3).
a, Representative immunofluorescence confocal imaging of P80 a2i/VPA/LIF-treated PSCCAG–GFP Fgfr2cnull and control WT + PSCCAG–GFP lungs double-labeled with GFP and airway differentiation markers multiciliated (β-tubulin4) and secretory (Scgb1a1) cells. Arrows: strong GFP overlapping signals in Fgfr2cnull mutants, contrasting with the less prominent signals (* asterisks) in WT; lower panels: single-channel images. b, Flexivent analysis of pulmonary function in CBC-complemented day 80 WT (n = 6) and Fgfr2cnull (n = 4); graph (mean ± s.e.m) showing non-significant (NS) difference in the resistance of conducting airways (Rn). c,d, Analysis of body weight in a2i/VPA/LIF PSCCAG–GFP complemented day 80 WT (n = 6), Fgfr2hetero (n = 3), Fgfr2cnull (n = 4) animals (c), and day 50 WT (n = 12), Ctnnb1hetero (n = 5), Ctnnb1cnull (n = 3) animals (d). Graph (mean ± s.e.m) indicating no significant difference in body weight between genotypes in both Fgfr2 and Ctnnb1 models. e, IF of day50 a2i/VPA/LIF-treated PSCCAG–GFP + Ctnnb1cnull lungs (n = 3). Representative image of GFP double labeling with markers of secretory (Scgb1a1) or basal (p63) cells in adult airways (arrows: depicts strong GFP staining contrasting). Statistical analyses: Student’s t-test (b), one-way ANOVA (c,d); NS: statistically non-significant: P > 0.05. Scale bars: a,e = 10 μm, 5 μm, respectively.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2, and Table 1
Source data
Source Data Fig. 1
Statistical Source Data of Fig. 1
Source Data Fig. 2
Statistical Source Data of Fig. 2
Source Data Fig. 3
Statistical Source Data of Fig. 3
Source Data Extended Data Fig. 2
Statistical Source Data of Extended Data Fig. 2
Source Data Extended Data Fig. 4
Statistical Source Data of Extended Data Fig. 4
Source Data Extended Data Fig. 5
Statistical Source Data of Extended Data Fig. 5
Source Data Extended Data Fig. 7
Statistical Source Data of Extended Data Fig. 7
Source Data Extended Data Fig. 8
Statistical Source Data of Extended Data Fig. 8
Source Data Extended Data Fig. 9
Statistical Source Data of Extended Data Fig. 9
Source Data Extended Data Fig. 9
Unprocessed western blots of Extended Data Fig. 9
Source Data Extended Data Fig. 10
Statistical Source Data of Extended Data Fig. 10
Rights and permissions
About this article
Cite this article
Mori, M., Furuhashi, K., Danielsson, J.A. et al. Generation of functional lungs via conditional blastocyst complementation using pluripotent stem cells. Nat Med 25, 1691–1698 (2019). https://doi.org/10.1038/s41591-019-0635-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-019-0635-8
This article is cited by
-
Replacing renal function using bioengineered tissues
Nature Reviews Bioengineering (2023)
-
An immunostimulatory glycolipid that blocks SARS-CoV-2, RSV, and influenza infections in vivo
Nature Communications (2023)
-
A novel renal perivascular mesenchymal cell subset gives rise to fibroblasts distinct from classic myofibroblasts
Scientific Reports (2022)
-
In vitro and in vivo functions of T cells produced in complemented thymi of chimeric mice generated by blastocyst complementation
Scientific Reports (2022)
-
Generation of inner ear sensory neurons using blastocyst complementation in a Neurog1+/−−deficient mouse
Stem Cell Research & Therapy (2021)