Abstract
Type 2 diabetes is characterized by insulin resistance and a gradual loss of pancreatic beta cell mass and function1,2. Currently, there are no therapies proven to prevent beta cell loss and some, namely insulin secretagogues, have been linked to accelerated beta cell failure, thereby limiting their use in type 2 diabetes3,4. The adipokine adipsin/complement factor D controls the alternative complement pathway and generation of complement component C3a, which acts to augment beta cell insulin secretion5. In contrast to other insulin secretagogues, we show that chronic replenishment of adipsin in diabetic db/db mice ameliorates hyperglycemia and increases insulin levels while preserving beta cells by blocking dedifferentiation and death. Mechanistically, we find that adipsin/C3a decreases the phosphatase Dusp26; forced expression of Dusp26 in beta cells decreases expression of core beta cell identity genes and sensitizes to cell death. In contrast, pharmacological inhibition of DUSP26 improves hyperglycemia in diabetic mice and protects human islet cells from cell death. Pertaining to human health, we show that higher concentrations of circulating adipsin are associated with a significantly lower risk of developing future diabetes among middle-aged adults after adjusting for body mass index (BMI). Collectively, these data suggest that adipsin/C3a and DUSP26-directed therapies may represent a novel approach to achieve beta cell health to treat and prevent type 2 diabetes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All requests for raw and analyzed data will be reviewed to verify if the request is subject to any intellectual property or confidentiality obligations. Any data and materials that can be shared will be released via a Material Transfer Agreement. Data from Framingham Heart Study participants are publicly available at dbGap according to NIH data sharing policies (study accession no. phs000007.v29.p10).
References
Alejandro, E. U., Gregg, B., Blandino-Rosano, M., Cras-Meneur, C. & Bernal-Mizrachi, E. Natural history of beta-cell adaptation and failure in type 2 diabetes. Mol. Aspects Med. 42, 19–41 (2015).
Rahier, J., Guiot, Y., Goebbels, R. M., Sempoux, C. & Henquin, J. C. Pancreatic beta-cell mass in European subjects with type 2 diabetes. Diabetes Obes. Metab. 10(Suppl. 4), 32–42 (2008).
Turner, R. C., Cull, C. A., Frighi, V. & Holman, R. R. Glycemic control with diet, sulfonylurea, metformin or insulin in patients with type 2 diabetes mellitus: progressive requirement for multiple therapies (UKPDS 49). UK Prospective Diabetes Study (UKPDS) Group. JAMA 281, 2005–2012 (1999).
Kahn, S. E. et al. Glycemic durability of rosiglitazone, metformin or glyburide monotherapy. N. Engl. J. Med 355, 2427–2443 (2006).
Lo, J. C. et al. Adipsin is an adipokine that improves beta cell function in diabetes. Cell 158, 41–53 (2014).
DeFronzo, R. A. et al. Type 2 diabetes mellitus. Nat. Rev. Dis. Primers 1, 15019 (2015).
Kahn, S. E., Cooper, M. E. & Del Prato, S. Pathophysiology and treatment of type 2 diabetes: perspectives on the past, present and future. Lancet 383, 1068–1083 (2014).
Kasuga, M. Insulin resistance and pancreatic beta cell failure. J. Clin. Invest. 116, 1756–1760 (2006).
Ferrannini, E. The stunned beta cell: a brief history. Cell Metab. 11, 349–352 (2010).
Ouchi, N., Parker, J. L., Lugus, J. J. & Walsh, K. Adipokines in inflammation and metabolic disease. Nat. Rev. Immunol. 11, 85–97 (2011).
Ilegems, E. et al. Reporter islets in the eye reveal the plasticity of the endocrine pancreas. Proc. Natl Acad. Sci. USA 110, 20581–20586 (2013).
Li, G. et al. Multifunctional in vivo imaging of pancreatic islets during diabetes development. J. Cell Sci. 129, 2865–2875 (2016).
Dalboge, L. S. et al. Characterisation of age-dependent beta cell dynamics in the male db/db mice. PLoS One 8, e82813 (2013).
Rosen, B. S. et al. Adipsin and complement factor D activity: an immune-related defect in obesity. Science 244, 1483–1487 (1989).
Gomez-Banoy, N. & Lo, J. C. Genetic manipulation with viral vectors to assess metabolism and adipose tissue function. Methods Mol. Biol. 1566, 109–124 (2017).
Baron, A. D., Schaeffer, L., Shragg, P. & Kolterman, O. G. Role of hyperglucagonemia in maintenance of increased rates of hepatic glucose output in type II diabetics. Diabetes 36, 274–283 (1987).
Mezza, T. et al. Insulin resistance alters islet morphology in nondiabetic humans. Diabetes 63, 994–1007 (2014).
Henquin, J. C. & Rahier, J. Pancreatic alpha cell mass in European subjects with type 2 diabetes. Diabetologia 54, 1720–1725 (2011).
Talchai, C., Xuan, S., Lin, H. V., Sussel, L. & Accili, D. Pancreatic beta cell dedifferentiation as a mechanism of diabetic beta cell failure. Cell 150, 1223–1234 (2012).
Guo, S. et al. Inactivation of specific beta cell transcription factors in type 2 diabetes. J. Clin. Invest. 123, 3305–3316 (2013).
Kim-Muller, J. Y. et al. Aldehyde dehydrogenase 1a3 defines a subset of failing pancreatic beta cells in diabetic mice. Nat. Commun. 7, 12631 (2016).
Dahan, T. et al. Pancreatic beta-cells express the fetal islet hormone gastrin in rodent and human. Diabetes 66, 426–436 (2017).
Sharma, R. B. & Alonso, L. C. Lipotoxicity in the pancreatic beta cell: not just survival and function, but proliferation as well? Curr. Diab. Rep. 14, 492 (2014).
Wang, J. Y., Lin, C. H., Yang, C. H., Tan, T. H. & Chen, Y. R. Biochemical and biological characterization of a neuroendocrine-associated phosphatase. J. Neurochem. 98, 89–101 (2006).
Shi, Y. et al. NSC-87877 inhibits DUSP26 function in neuroblastoma resulting in p53-mediated apoptosis. Cell Death Dis. 6, e1841 (2015).
Song, M. et al. NSC-87877, inhibitor of SHP-1/2 PTPs, inhibits dual-specificity phosphatase 26 (DUSP26). Biochem. Biophys. Res. Commun. 381, 491–495 (2009).
Belgardt, B. F. et al. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat. Med. 21, 619–627 (2015).
Prentki, M. & Nolan, C. J. Islet beta cell failure in type 2 diabetes. J. Clin. Invest. 116, 1802–1812 (2006).
Zhou, Q et al. Relationship between serum adipsin and the first phase of glucose-stimulated insulin secretion in individuals with different glucose tolerance. J. Diabetes Investig. 9, 1128–1134 (2018).
Rosen, E. D. & Spiegelman, B. M. What we talk about when we talk about fat. Cell 156, 20–44 (2014).
Type 2 Diabetes Knowledge Portal rs2930902. type2diabetesgenetics.org 2019 Feb 1; http://www.type2diabetesgenetics.org/variantInfo/variantInfo/rs2930902#
Type 2 Diabetes Knowledge Portal rs2930900. type2diabetesgenetics.org 2019 Feb 1; http://www.type2diabetesgenetics.org/variantInfo/variantInfo/rs2930900#
Type 2 Diabetes Knowledge Portal rs7257521. type2diabetesgenetics.org 2019 Feb 1; http://www.type2diabetesgenetics.org/variantInfo/variantInfo/rs7257521#
Type 2 Diabetes Knowledge Portal rs17684161. type2diabetesgenetics.org 2019 Feb 1; http://www.type2diabetesgenetics.org/variantInfo/variantInfo/rs17684161#
Wang, T. J. et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 17, 448–453 (2011).
Kettunen, J. et al. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat. Commun. 7, 11122 (2016).
Ishida, E., Kim-Muller, J. Y. & Accili, D. Pair feeding, but not insulin, phloridzin or rosiglitazone treatment, curtails markers of beta-cell dedifferentiation in db/db mice. Diabetes 66, 2092–2101 (2017).
Camporez, J. P. et al. Cellular mechanism by which estradiol protects female ovariectomized mice from high-fat diet-induced hepatic and muscle insulin resistance. Endocrinology 154, 1021–1028 (2013).
Mellado-Gil, J. et al. Disruption of hepatocyte growth factor/c-Met signaling enhances pancreatic beta-cell death and accelerates the onset of diabetes. Diabetes 60, 525–536 (2011).
Burns, S. M. et al. High-throughput luminescent reporter of insulin secretion for discovering regulators of pancreatic beta-cell function. Cell Metab. 21, 126–137 (2015).
Rappsilber, J., Ishihama, Y. & Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 (2003).
Elias, J. E. & Gygi, S. P. Target–decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 (2007).
Huttlin, E. L. et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 (2010).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Pelossof, R. et al. Prediction of potent shRNAs with a sequential classification algorithm. Nat. Biotechnol. 35, 350–353 (2017).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Soneson, C., Love, M. I. & Robinson, M. D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 4, 1521 (2015).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108 (2013).
Splansky, G. L. et al. The third generation cohort of the national heart, lung and blood institute’s Framingham Heart Study: design, recruitment and initial examination. Am. J. Epidemiol. 165, 1328–1335 (2007).
Kannel, W. B., Feinleib, M., McNamara, P. M., Garrison, R. J. & Castelli, W. P. An investigation of coronary heart disease in families. The Framingham offspring study. Am. J. Epidemiol. 110, 281–290 (1979).
Carson, R. T. & Vignali, D. A. Simultaneous quantitation of 15 cytokines using a multiplexed flow cytometric assay. J. Immunol. Methods 227, 41–52 (1999).
dupont, N. C., Wang, K., Wadhwa, P. D., Culhane, J. F. & Nelson, E. L. Validation and comparison of luminex multiplex cytokine analysis kits with ELISA: determinations of a panel of nine cytokines in clinical sample culture supernatants. J. Reprod. Immunol. 66, 175–191 (2005).
Matthews, D. R. et al. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28, 412–419 (1985).
Lee, J. J. et al. Cross-sectional associations of computed tomography (CT)-derived adipose tissue density and adipokines: the Framingham Heart Study. J. Am. Heart Assoc. 5, e002545 (2016).
Sotoodehnia, N. et al. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat. Genet. 42, 1068–1076 (2010).
Staley, J. R. et al. PhenoScanner: a database of human genotype–phenotype associations. Bioinformatics 32, 3207–3209 (2016).
Consortium, G. T. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Acknowledgements
N.G.-B. is supported by an American Diabetes Association postdoctoral fellowship (1-18-PMF-032). J.S.G. was supported by an MGH NIH T32 Training Grant (HL007208), the John S. LaDue Memorial Fellowship and the MGH Physician Scientist Development Program. This work was supported by a Weill Cornell Department of Medicine Seed Grant for Innovative Research to J.C.L., the JPB Foundation (to B.M.S.), Jill Roberts IBD Institute (to G.P.) and NIH grants DK097303 (to J.C.L.), R03 DK111762 (to J.C.L.), R01 DK121844 (to J.C.L.), R01 HL140224 (to J.E.H.) and R01 HL134893 (to J.E.H.). This work was partially supported by the National Heart, Lung and Blood Institute’s Framingham Heart Study (contracts N01-HC-25195 and HHSN268201500001I) and by the Division of Intramural Research (to P.C., G.S., C.L., S.-J.H. and D.L.) of the National Heart, Lung and Blood Institute. We acknowledge support from the Yale Mouse Metabolic Phenotyping Center via NIH grants nos. U24 DK-059635, R01 DK116774, R01 DK114793 and P30 DK045735 (all to G.I.S.). We acknowledge the Microscopy and Image Analysis Core Facility at Weill Cornell Medicine for analysis of the images presented in this study, J. Cao for his help with confocal microscopy and the Human Islet and Adenovirus Core of the Einstein–Sinai Diabetes Research Center (NIH grant no. P30 DK020541-38) for the human islet studies. The views expressed in this Letter are those of the authors and do not necessarily represent the views of the National Institute of Diabetes and Digestive and Kidney Diseases, National Heart, Lung and Blood Institute, the National Institutes of Health or the US Department of Health and Human Services.
Author information
Authors and Affiliations
Contributions
J.C.L. and N.G.-B. designed the animal, cellular and molecular studies. J.E.H. and J.S.G. designed the human study. N.G.-B., G.L., A.R.-N., T.C., B.P., M.A.P., J.P.C. and R.J.P. performed and analyzed the animal experiments. N.G.-B, A.R.-N., T.C., B.P. and C.R. developed the in vitro experiments. J.E.H., J.S.G., V.B., S.-J.H., C.Y., D.L. and M.G.L. analyzed the human study data. S.M. analyzed microscope images. N.D. developed and analyzed proteomics experiments. G.P. analyzed RNA sequencing experiments. L.E.D., G.I.S., A.G.-O., M.H. and B.M.S. provided scientific input and analyzed the data. J.C.L., J.E.H., N.G.-B. and J.S.G. wrote the manuscript, and all authors contributed to writing and provided feedback. J.C.H. (jho1@mgh.harvard.edu) is the contact for the Framingham studies.
Corresponding authors
Ethics declarations
Competing interests
Cornell University has filed a provisional patent application that covers the use of DUSP26 inhibitors for the treatment of type 2 diabetes. (US patent application no. 62/740744; N.G.-B. and J.C.L.). V.B. is currently an employee of Fractyl Laboratories Inc. and all analyses were conducted during employment at Massachusetts General Hospital.
Additional information
Peer review information Joao Monteiro was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
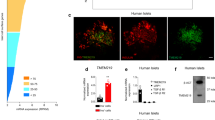
Extended Data Fig. 1 Islet area in the eye is correlated with beta cell area in the pancreas and lower fasting glucose.
(a) Scatterplot of fasting glucose levels and eye islet area in db/db mice transplanted with islets in the anterior chamber of the eye and treated with AAV-Adipsin or AAV GFP (n = 5 per group). (b) Scatterplot of beta cell area and eye islet area in db/db mice transplanted with islets in the anterior chamber of the eye and treated with AAV-Adipsin or AAV-GFP (n = 5 per group). (c) Representative immunohistochemistry images of insulin (brown) performed in pancreatic sections from db/db-GFP and db/db-Adipsin mice. IHC was performed at least twice independently with similar results. Scale bars, 100 µm. (d) Quantification of beta cell area in db/db mice transplanted with islets in the anterior chamber of the eye and treated with AAV-Adipsin or AAV-GFP (n = 5 per group). Data were analyzed by two-tailed unpaired t-test. (p = 0.027). (e) Western blot for adipsin from sera of B6 WT mice injected with GFP-AAV or adipsin-AAV after 5 months (n = 4 per group) along with quantification. (f) Quantification of islet area in the eye over time in WT-GFP and WT-Adipsin mice (n = 5 WT-Adipsin, n = 6 WT-GFP). Eye imaging was repeated at least 3 times independently with similar results. (g) Reporter islets transplanted into the anterior chamber of the eyes of WT-GFP and WT-Adipsin mice were serially imaged by light microscopy. Representative images are from a single mouse in each treatment group at the indicated time points. Islets are outlined within dashed white circles. Islets are amplified inside the white box. Eye imaging was repeated at least 3 times independently with similar results. Scale bars, 1 mm. Data are expressed as mean ± s.e.m. *P < 0.05, ** P < 0.01, *** P < 0.001.
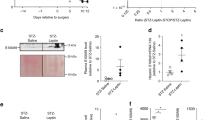
Extended Data Fig. 2 Treatment with AAV-Adipsin prevents beta cell failure without affecting body weight or insulin sensitivity.
(a) Cfd mRNA levels were quantified by qPCR in the indicated tissues from db/db mice transduced with AAV-Adipsin or AAV-GFP at 1 month post-transduction (iWAT: n = 3 db/db-Adipsin, n = 4 db/db-GFP. vWAT: n = 3 for both groups. Liver: n = 4 for both groups. Kidney: n = 6 db/db-Adipsin, n = 5 db/db GFP. Islets: n = 2 for both groups). (b) Mouse serum adipsin levels were measured by ELISA (n = 5 db/db-GFP, n = 5 db/db-Adipsin, n = 2 B6 WT, n = 3 Adipsin KO). (c) Body weights of db/db mice transduced with AAV-Adipsin versus AAV-GFP at the indicated time points (n = 8 mice per group at 1 month post transduction and n = 15-16 mice per group at 1.5-months post transduction). (d) Glucose infusion rate during hyperinsulinemic euglycemic clamp in db/db mice treated with adipsin versus controls (n = 5 db/db-Adipsin, n = 6 db/db-GFP). (e) Scatterplot of fasting glucose and beta cell mass in db/db mice transduced with AAV-Adipsin versus controls (n = 18-21 per group). (f) Scatterplot of fasting insulin and beta cell mass in db/db mice injected with AAV-Adipsin versus controls (n = 18-21 per group). (g) Representative IHC staining for insulin (brown) in pancreases of two control and two adipsin transduced db/db mice. Areas in the dashed boxes are amplified in the panels at the right of each image. IHC was repeated at least twice independently with similar results. Scale bars, 1 mm left panels, 100 µm amplified regions. Data are expressed as mean ± s.e.m.
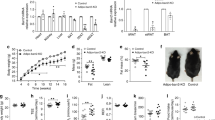
Extended Data Fig. 3 Adipsin prevents beta cell death and prevents loss of beta cell transcriptional identity.
(a) Representative images of immunofluorescence (IF) staining for insulin and TUNEL assay in pancreases of db/db mice injected with AAV-Adipsin versus controls. White dashed box indicates region magnified in white panel. IF was repeated at least twice independently with similar results. (b) Quantification of TUNEL + beta cells as determined by IF. (n = 4 mice per group at 1.5 months, n = 5 mice per group at 6 months). Data were analyzed by two-tailed unpaired t-test. (p = 0.03). (c) Representative images of immunofluorescence (IF) staining for insulin and Ki67 in pancreases of the indicated groups of mice. White dashed box indicates region magnified in white panel. IF was repeated at least twice independently with similar results. (d) Quantification of Ki67 + beta cells as determined by IF (At 1.5 months; n = 6 mice per group. At 6 months n = 3 db/db-Adipsin, n = 4 db/db-GFP). (e) Glucagon positive cells were quantitated by IHC in the pancreases of db/db mice transduced with AAV-Adipsin versus controls. Representative images are shown. IHC was repeated at least three times independently with similar results. (f) Quantification of alpha cell mass in B6 WT mice, db/db mice treated with AAV-GFP and db/db mice treated with adipsin AAV at the indicated time points (Pre AAV; n = 4 db/db, n = 4 B6 WT. At 1.5 months post-AAV; n = 6 db/db-Adipsin, n = 5 db/db-GFP, n = 2 B6 WT. At 6 months post-AAV; n = 13 db/db-Adipsin, n = 16 db/db-GFP, n = 4 B6 WT). Data were analyzed by two-tailed unpaired t-test. (At 1.5 months post-AAV db/db-GFP vs db/db-Adipsin p = 0.03). (g) Quantification of the ratio between alpha cell mass and beta cell mass in pancreases from the specified groups at the indicated time points (Pre AAV; n = 4 db/db, n = 4 B6 WT. At 1.5 months post-AAV; n = 6 db/db-Adipsin, n = 5 db/db-GFP, n = 2 B6 WT. At 6 months post-AAV; n = 13 db/db-Adipsin, n = 16 db/db-GFP, n = 4 B6 WT). Data were analyzed by two-tailed unpaired t-test. (At 6 months post-AAV db/db-GFP vs db/db-Adipsin p = 0.029). (h) Fasting serum glucagon levels in db/db mice transduced with AAV-GFP or AAV-Adipsin (n = 8 per group). Scale bars, 100 µm (a, c and e). Data are expressed as mean ± s.e.m. *P < 0.05, ** P < 0.01, *** P < 0.001.
Extended Data Fig. 4 Adipsin increases the expression of beta cell transcription factors and decreases gastrin expression.
(a,b) Representative images (a) of MAFA and insulin IF in B6 WT mice, db/db mice injected with AAV-Adipsin or AAV-GFP along with quantification (b) of MAFA + beta cells (n = 4 mice for B6 WT, n = 6 mice per db/db treatment group). Data were analyzed by two-tailed unpaired t-test. (p = 0.029). (c,d) Representative images (c) of NKX2-2 and insulin IF in the indicated groups along with quantification (d) of NKX2-2 + beta cells (n = 5 mice per group). Data were analyzed by two-tailed unpaired t-test. (p = 0.027). (e,f) Representative images (e) of gastrin and insulin IF in the indicated groups along with quantification (f) of gastrin + beta cells (n = 5 mice per group). White dashed box indicates region magnified in white panel. Data were analyzed by two-tailed unpaired t-test. (p = 0.038). Scale bars, 100 µm (a, b and c). Data are expressed as mean ± s.e.m. *P < 0.05, ** P < 0.01, *** P < 0.001.
Extended Data Fig. 5 Adipsin/C3a increases insulin secretion and protects from beta cell death by inhibiting DUSP26.
(a) Ins1 beta cells were subjected to a glucose-stimulated insulin secretion assay at 0 or 17 mM glucose with or without C3a (n = 8 per group). Results are representative of three independent experiments. Data were analyzed by two-tailed unpaired t-test. (Glc 0 mM vs glc 17 mM p = 0.0019, glc 17 mM vs glc 17 mM + C3a p = 0.009). (b) Representative gating strategy for islet cells. (c) Heatmap of genes significantly changed by palmitate treatment and counter-regulated by C3a in WT islets (n = 3 per group) Colors show raw z-scores of mean normalized counts. (d) GO biological process analysis of genes whose expression were downregulated by palmitate and counter-regulated (increased) by C3a. Data were analyzed by Fisher/binominal test with Bonferroni-adjusted P value (n = 33 genes) (e) GO biological process analysis of genes whose expression were upregulated by palmitate and counter-regulated (decreased) by C3a. Data were analyzed by Fisher/binominal test with Bonferroni-adjusted P value (n = 43 genes) (f) Pathway analysis from the DEPOD database depicting significant phosphatase substrates in islets that are regulated at the gene expression level by C3a from Fig. 3d. (n = 76 genes) Data were analyzed by Fisher/binominal test with Bonferroni-adjusted P value (g) Ins2 and Gcg were determined by qPCR in isolated pancreatic islets from WT mice transduced with Dusp26 or control lentivirus (n = 4 GFP, n = 3 Dusp26). Data are representative of 3 independent experiments. Data were analyzed by two-tailed unpaired t-test. (Ins2 p = 0.0284, Gcg p = 0.033). (h) Cell viability was determined in INS-1 cells transduced with Dusp26 and controls (n = 6 per group). Data are representative of 3 independent experiments. Data were analyzed by two-tailed unpaired t-test. (GFP in veh vs GFP in Pa p = 0.022, Dusp26 in veh vs Dusp26 in Pa p = 0.0000047, GFP in Pa vs Dusp26 in Pa p = 0.0004). (i) Representative western blot analysis of DUSP26 in INS-1 cells treated with shRNA against Dusp26 versus control shRNA. Data are representative of 3 independent experiments. (j) Quantification of DUSP26 expression for Supplementary Figure 8d from 3 independent experiments. Data were analyzed by two-tailed unpaired t-test. (p = 0.028). (k) Body weight in db/db mice treated for 2 weeks with NSC-87877 and controls at the indicated time points (n = 15 NSC-87877 group, n = 16 saline group). (l) Representative western blot analysis for FLAG after immunoprecipitation of the DUSP26 protein complex using FLAG-M2 antibodies in lysates of INS-1 cells overexpressing control GFP, intact DUSP26 (D26) and catalytically inactive DUSP26 mutant (C152S). Data are representative of at least 3 independent experiments. (m) KEGG pathway analysis of proteins enriched in DUSP26 immunoprecipitation experiment. Proteins with log2 ≥ 1.5 between control GFP and DUSP26 mutant (C152S) were included in the analysis. Top and selected pathways are shown. Data were analyzed by Fisher/binominal test with Bonferroni-adjusted P value (n = 253 genes) (n) Heatmap of selected proteins belonging to relevant pathways and significantly enriched in the DUSP26 mutant (C152S) pulldown. (n = 2 per group). Colors show log2 fractional intensity. Data are expressed as mean ± s.e.m. *P < 0.05, ** P < 0.01, *** P < 0.001.
Extended Data Fig. 6 Baseline participant characteristics.
The average adipsin concentration was 900 ± 273 ng/mL (mean ± s.d.) Values depict mean ± standard deviation where appropriate.
Extended Data Fig. 7 Clinical correlates of adipsin.
(a) In cross-section multivariable analyses, higher adipsin levels associate with lower odds of diabetes (OR 0.69) and with higher odds of obesity (OR 1.68). Adipsin is associated with a number of clinical traits including BMI, waist circumference, diastolic blood pressure reduction and reduced HDL cholesterol. Additionally, higher levels associate with significantly lower fasting glucose, with a trend toward improved insulin resistance as measured by HOMA-IR that did not meet the Bonferroni-corrected p-value threshold for significance. Analyses were multivariable linear (t statistic) or logistic regression models (Wald Chi-square test) with two-sided p-values deemed significant at a Bonferroni-corrected p-value threshold of p = 0.05/10 primary traits = 0.005. Similar results were obtained in secondary analyses stratified by Framingham cohort (Offspring vs. Third Generation). (b) Cross-sectional radiographic anatomically specific adiposity volumes were obtained from participants of the Third-Generation Framingham cohort (n = 3068, 2002 to 2005). In secondary analyses, volumetric measures of adiposity reveal that adipsin is strongly associated with subcutaneous (SAT) and intrathoracic adipose volumes and not visceral adipose volumes. With regard to subcutaneous adipose, a one standard deviation rise in adipsin was associated with a 0.4 standard deviation increase in subcutaneous adipose volume. Analyses were multivariable linear regression models (t statistic) with two-sided p-values deemed significant at a p-value threshold of p = 0.05.
Extended Data Fig. 8 GWAS of adipsin levels in the Framingham Cohort.
A genome-wide association study of adipsin levels was conducted among n = 6791 individuals with available genetic data using an additive genetic model (t statistic). (a) A quintile-quintile plot of the observed and expected P-values. Each dot represents observed data while the solid line represents the null hypothesis of no association. (b) A regional plot of the sentinel SNP (rs2930902) associated with elevated plasma adipsin level is housed intronically within the MED16. Other significant cis-SNPs are also shown.
Extended Data Fig. 9 Significant cis variant SNPs with phenotypic association, eQTL analysis, and associated metabolites from published databases.
The lead SNP on chromosome 19 rs2930902 is an intron variant in the MED16 locus and is associated with type 2 diabetes. Tissue specific cis-eQTL’s reveal that this SNP is significantly associated with adipsin expression in a number of tissues but notably in subcutaneous (but not visceral) adipose tissue. Moreover, the same SNP is associated with known branch chain amino acid metabolites (glycine and valine) known to predict incident type 2 diabetes mellitus. Three additional SNPs associated with adipisin expression are shown.
Supplementary information
Supplementary Information
Supplementary Table 1
Rights and permissions
About this article
Cite this article
Gómez-Banoy, N., Guseh, J.S., Li, G. et al. Adipsin preserves beta cells in diabetic mice and associates with protection from type 2 diabetes in humans. Nat Med 25, 1739–1747 (2019). https://doi.org/10.1038/s41591-019-0610-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-019-0610-4
This article is cited by
-
Plasma proteome profiling reveals the therapeutic effects of the PPAR pan-agonist chiglitazar on insulin sensitivity, lipid metabolism, and inflammation in type 2 diabetes
Scientific Reports (2024)
-
Adipsin alleviates cardiac microvascular injury in diabetic cardiomyopathy through Csk-dependent signaling mechanism
BMC Medicine (2023)
-
Alterations of adipokines, pancreatic hormones and incretins in acute and convalescent COVID-19 children
BMC Pediatrics (2023)
-
A beta cell subset with enhanced insulin secretion and glucose metabolism is reduced in type 2 diabetes
Nature Cell Biology (2023)
-
Plasma and aqueous levels of alarin and adipsin in patients with and without diabetic retinopathy
BMC Ophthalmology (2022)