Abstract
Fanconi anemia (FA) is a DNA repair syndrome generated by mutations in any of the 22 FA genes discovered to date1,2. Mutations in FANCA account for more than 60% of FA cases worldwide3,4. Clinically, FA is associated with congenital abnormalities and cancer predisposition. However, bone marrow failure is the primary pathological feature of FA that becomes evident in 70–80% of patients with FA during the first decade of life5,6. In this clinical study (ClinicalTrials.gov, NCT03157804; European Clinical Trials Database, 2011-006100-12), we demonstrate that lentiviral-mediated hematopoietic gene therapy reproducibly confers engraftment and proliferation advantages of gene-corrected hematopoietic stem cells (HSCs) in non-conditioned patients with FA subtype A. Insertion-site analyses revealed the multipotent nature of corrected HSCs and showed that the repopulation advantage of these cells was not due to genotoxic integrations of the therapeutic provirus. Phenotypic correction of blood and bone marrow cells was shown by the acquired resistance of hematopoietic progenitors and T lymphocytes to DNA cross-linking agents. Additionally, an arrest of bone marrow failure progression was observed in patients with the highest levels of gene marking. The progressive engraftment of corrected HSCs in non-conditioned patients with FA supports that gene therapy should constitute an innovative low-toxicity therapeutic option for this life-threatening disorder.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its Extended Data and Supplementary Information files.
References
Bagby, G. Recent advances in understanding hematopoiesis in Fanconi anemia. F1000Res. 7, 105 (2018).
Knies, K. et al. Biallelic mutations in the ubiquitin ligase RFWD3 cause Fanconi anemia. J. Clin. Invest. 127, 3013–3027 (2017).
Casado, J. A. et al. A comprehensive strategy for the subtyping of Fanconi anemia patients: conclusions from the Spanish Fanconi Anemia research network. J. Med. Genet. 44, 241–249 (2007).
Taniguchi, T. & D’Andrea, A. D. Molecular pathogenesis of Fanconi anemia: recent progress. Blood 107, 4223–4233 (2006).
Butturini, A. et al. Hematologic abnormalities in Fanconi anemia: an International Fanconi Anemia Registry study. Blood 84, 1650–1655 (1994).
Kutler, D. I. et al. A 20-year perspective on the International Fanconi Anemia Registry (IFAR). Blood 101, 1249–1256 (2003).
Ceccaldi, R. et al. Bone marrow failure in Fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell 11, 36–49 (2012).
Croop, J. M. et al. Mobilization and collection of peripheral blood CD34+ cells from patients with Fanconi anemia. Blood 98, 2917–2921 (2001).
Kelly, P. F. et al. Stem cell collection and gene transfer in Fanconi anemia. Mol. Ther. 15, 211–219 (2007).
Liu, J. M. et al. Engraftment of hematopoietic progenitor cells transduced with the Fanconi anemia group C gene (FANCC). Hum. Gene Ther. 10, 2337–2346 (1999).
Gross, M. et al. Reverse mosaicism in Fanconi anemia: natural gene therapy via molecular self-correction. Cytogenet. Genome Res. 98, 126–135 (2002).
Hamanoue, S. et al. Myeloid lineage-selective growth of revertant cells in Fanconi anaemia. Br. J. Haematol. 132, 630–635 (2006).
Mankad, A. et al. Natural gene therapy in monozygotic twins with Fanconi anemia. Blood 107, 3084–3090 (2006).
Soulier, J. et al. Detection of somatic mosaicism and classification of Fanconi anemia patients by analysis of the FA/BRCA pathway. Blood 105, 1329–1336 (2005).
Adair, J. E. et al. Lessons learned from two decades of clinical trial experience in gene therapy for Fanconi anemia. Curr. Gene Ther. 16, 338–348 (2017).
Rio, P. et al. Engraftment and in vivo proliferation advantage of gene-corrected mobilized CD34+ cells from Fanconi anemia patients. Blood 130, 1535–1542 (2017).
Meyer, S., Neitzel, H. & Tonnies, H. Chromosomal aberrations associated with clonal evolution and leukemic transformation in Fanconi anemia: clinical and biological implications. Anemia 2012, 349837 (2012).
Quentin, S. et al. Myelodysplasia and leukemia of Fanconi anemia are associated with a specific pattern of genomic abnormalities that includes cryptic RUNX1/AML1 lesions. Blood 117, e161–e170 (2011).
Aiuti, A. et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott–Aldrich syndrome. Science 341, 1233151 (2013).
Biffi, A. et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 341, 1233158 (2013).
Cartier, N. et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science 326, 818–823 (2009).
Eichler, F. et al. Hematopoietic stem-cell gene therapy for cerebral adrenoleukodystrophy. N. Engl. J. Med. 377, 1630–1638 (2017).
Thompson, A. A. et al. Gene therapy in patients with transfusion-dependent β-thalassemia. N. Engl. J. Med. 378, 1479–1493 (2018).
Molina-Estevez, F. J. et al. Lentiviral-mediated gene therapy in Fanconi anemia-A mice reveals long-term engraftment and continuous turnover of corrected HSCs. Curr. Gene Ther. 15, 550–562 (2015).
Lex, A., Gehlenborg, N., Strobelt, H., Vuillemot, R. & Pfister, H. UpSet: visualization of intersecting sets. IEEE Trans. Vis. Comput. Graph. 20, 1983–1992 (2014).
Battaile, K. P. et al. In vivo selection of wild-type hematopoietic stem cells in a murine model of Fanconi anemia. Blood 94, 2151–2158 (1999).
Galimi, F. et al. Gene therapy of Fanconi anemia: preclinical efficacy using lentiviral vectors. Blood 100, 2732–2736 (2002).
Gush, K. A., Fu, K. L., Grompe, M. & Walsh, C. E. Phenotypic correction of Fanconi anemia group C knockout mice. Blood 95, 700–704 (2000).
Haneline, L. S. et al. Retroviral-mediated expression of recombinant Fancc enhances the repopulating ability of Fancc −/− hematopoietic stem cells and decreases the risk of clonal evolution. Blood 101, 1299–1307 (2003).
Muller, L. U. et al. Rapid lentiviral transduction preserves the engraftment potential of Fanca −/− hematopoietic stem cells. Mol. Ther. 16, 1154–1160 (2008).
Rio, P. et al. In vitro phenotypic correction of hematopoietic progenitors from Fanconi anemia group A knockout mice. Blood 100, 2032–2039 (2002).
Alter, B. P., Giri, N., Savage, S. A. & Rosenberg, P. S. Cancer in the National Cancer Institute Inherited Bone Marrow Failure Syndrome cohort after fifteen years of follow-up. Haematologica 103, 30–39 (2018).
Masserot, C. et al. Head and neck squamous cell carcinoma in 13 patients with Fanconi anemia after hematopoietic stem cell transplantation. Cancer 113, 3315–3322 (2008).
Gonzalez-Murillo, A. et al. Development of lentiviral vectors with optimized transcriptional activity for the gene therapy of patients with Fanconi anemia. Hum. Gene Ther. 21, 623–630 (2010).
Castella, M. et al. Chromosome fragility in patients with Fanconi anaemia: diagnostic implications and clinical impact. J. Med. Genet. 48, 242–250 (2011).
Charrier, S. et al. Quantification of lentiviral vector copy numbers in individual hematopoietic colony-forming cells shows vector dose-dependent effects on the frequency and level of transduction. Gene Ther. 18, 479–487 (2011).
Schmidt, M. et al. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR). Nat. Methods 4, 1051–1057 (2007).
Gabriel, R. et al. Comprehensive genomic access to vector integration in clinical gene therapy. Nat. Med. 15, 1431–1436 (2009).
Paruzynski, A. et al. Genome-wide high-throughput integrome analyses by nrLAM-PCR and next-generation sequencing. Nat. Protoc. 5, 1379–1395 (2010).
Afzal, S., Wilkening, S., Von Kalle, C., Schmidt, M. & Fronza, R. GENE-IS: time-efficient and accurate analysis of viral integration events in large-scale gene therapy data. Mol. Ther. Nucleic Acids 6, 133–139 (2017).
Acknowledgements
The authors thank F. Mavilio, A. Thrasher, C. Booth and M. Cavazzana for collaboration in the EUROFANCOLEN Consortium, and D. A. Williams, H. Hanenberg, J. Tolar, G. P. Rizzardi and G. Shah for helpful discussions. The authors also thank A. de la Cal for coordinating the delivery of BM and PB samples from patients with FA. The authors are also indebted to the patients with FA, their families and clinicians from the Fundación Anemia de Fanconi. This work was supported by grants from the European Commission’s Seventh Framework Program (HEALTH-F5-2012-305421 to the EUROFANCOLEN Consortium J.B., J.Surrallés, C.D.H., J.Soulier, M.S., A.G. and J.Sevilla), Ministerio de Sanidad, Servicios Sociales e Igualdad (EC11/060 and EC11/550 to C.D.H., J.Sevilla, J.B. and J.Surrallés), Ministerio de Economía, Comercio y Competitividad and Fondo Europeo de Desarrollo Regional (SAF2015-68073-R and RTI2018-097125-B-I00 to P.R. and G.G.) and Fondo de Investigaciones Sanitarias at the Instituto de Salud Carlos III (RD12/0019/0023 to J.C.S.). The authors also thank the Fundación Botín for promoting translational research at the Hematopoietic Innovative Therapies division of the CIEMAT. CIBERER is an initiative of the Instituto de Salud Carlos III and Fondo Europeo de Desarrollo Regional. J.Soulier is supported by the ERC Candidator grant CEVAL-311660, and J.Surrallés is supported by ICREA Academia and SAF2015-64152-R.
Author information
Authors and Affiliations
Contributions
P.R., S.N., W.W., R.S.-D., R.M.P., J.C.S., M.B., E.M., N.W., R.S., M.L.Lamana, R.M.Y., J.A.C., Y.G., F.J.R.-R., L.A., O.A., A.R., G.G., M.L.Lozano., L.C., M.H., E.G., I.G., J.B., C.D.H., A.G., J.Surrallés, J.Soulier and M.S. performed the experimental studies and provided critical materials and reagents. R.H., N.G.A., R.L. and A.C. diagnosed patients and included them in the clinical trial. C.D.H. and J.Sevilla conducted the clinical trial. P.R., S.N., J.Sevilla and J.A.B. wrote the manuscript. J.D.S. reviewed the manuscript. P.R., S.N., C.D.H., J.Sevilla and J.A.B. designed the study. All authors discussed the results and contributed to the preparation of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The Hematopoietic Innovative Therapies division at CIEMAT receives funding and has licensed the PGK-FANCA-WPRE* lentiviral vector to Rocket Pharmaceuticals. J.A.B., J.Sevilla and J.C.S. are consultants for Rocket Pharmaceuticals. J.D.S. is Medical Director of Rocket Pharmaceuticals. P.R., S.N., J.A.C., J.C.S., J.Sevilla, G.G. and J.A.B. are inventors on patents on lentiviral vectors filed by CIEMAT, CIBERER and Fundación Jiménez Díaz, and may be entitled to receive financial benefits from the licensing of such patents. M.S. is co-founder and CEO of GeneWerk. W.W. is an employee of GeneWerk. The remaining authors declare no competing interests.
Additional information
Peer review information: Kate Gao was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended Data
Extended Data Fig. 1 Characteristics of the patients with FA enrolled in the gene therapy trial.
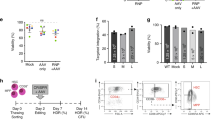
a,b, Genotype (a) and hematological and hematopoietic characteristics (b) of the recruited patients. In two patients (FA-02002 and FA-02004), CD34+ cells were collected and then cryopreserved until PB cell counts decreased to levels defined in the gene therapy trial. At that time, cells were thawed, transduced and infused. PB cell counts corresponding to patients FA-02005 and FA-02006 fit the criteria required for both the collection and the infusion of transduced cells. In these two patients, cells were transduced and infused immediately after collection. The table shows PB cell counts and the total content of hematopoietic progenitors per microliter of BM. Additionally, CFC resistance to MMC and percentages of T cells with chromosomal breaks after DEB challenge are shown. CFC values represent mean values corresponding to the scoring of three plates seeded with BM cells obtained before HSC collection and before HSC gene therapy. A total of 25–50 metaphases were analyzed in the DEB tests.
Extended Data Fig. 2 Characteristics of cell products transduced with the therapeutic PGK-FANCA-WPRE* lentiviral vector and infused in non-conditioned patients with FA-A.
G-CSF and plerixafor mobilized CD34+ cells were pre-stimulated for 8–10 h and then transduced with the therapeutic lentiviral vector for another 12–14 h, as described in the Methods. Values of VCN cell−1 were determined in hematopoietic colonies after 14 d of growth in semisolid cultures. In this case, determinations were carried out either in pooled (marked by a hashtag) or individual colonies (marked by a dollar sign). In no instance were mean VCN cell−1 values in lentiviral vector-positive colonies higher than 1.0 copies cell−1. Therefore analyses of the proportion of corrected cells in the PB and BM of treated patients (Fig. 1) are based on the assumption of the presence of 1.0 VCN in transduced cells. Cell viabilities in the infused products, including cryopreserved products, ranged between 84 and 97%. An asterisk denotes that CD34+ cells from one of the two apheresis procedures were purified using a modified immunoselection procedure used to improve the yield of CD34+ cells (see Methods). CFC values represent means ± s.e.m., corresponding to the scoring of three plates seeded with cells from each transduced product.
Extended Data Fig. 3 Representative cytogenetic analysis of BM cells from gene therapy-treated patients with FA-A.
a,b, Representative G-banding karyotypes (a) and FISH analyses of chromosomes 1q, 3q and 7q (b) of BM samples obtained before and after infusion of transduced cells. For cytogenetic studies, 10–20 metaphases of BM cells obtained before and at different time points after the infusion of transduced cells were examined. In total, 200 interphase nuclei per FISH were scored.
Extended Data Fig. 4 Cytogenetic and high-throughput sequencing studies in BM cells from patients with FA-A treated by gene therapy.
The table shows G-banding karyotypes, FISH analyses of chromosomes 1q, 3q and 7q, and array CGH analyses using 400K arrays (Agilent Technologies) of BM samples obtained before and at different times after infusion of transduced cells. The table shows that no cytogenetic nor genomic abnormalities were observed in any of the treated patients, either before or after the infusion of transduced HSCs. Cytogenetic studies were conducted in 10–20 metaphases. In total, 200 interphase nuclei per FISH were scored.
Extended Data Fig. 5 Genome-wide mapping of vector UISs in PB cells of the patients FA-02004 and FA-02005 after gene therapy.
The ten most represented UISs in PB cells from the patients FA-02004 and FA-02005 at different times after gene therapy are shown. Samples were analyzed as in Fig. 3. UISs detected at more than one time point, or with a contribution higher than 1% at one time point are marked with individualized colors. UISs marked in dark gray are those not represented in the top ten rank. RefSeq genes nearest to the UISs are listed in the table. The top ten clones marked in light gray indicate clones with a contribution lower than 1%. The total number of UISs is indicated at the bottom of each column. Because of the high contribution of clone B3GNT3 in PB cells from patient FA-02004 at 18 months post-infusion, analyses carried out at 24 months were conducted in sorted cell lineages, which revealed a marked reduction in the representation of this clone.
Extended Data Fig. 6 Hematopoietic progenitor content in patients with FA-A before and after gene therapy.
Analysis of the CFC content in BM cells from patients with FA before and after infusion of transduced CD34+ cells. Numbers of hematopoietic colonies generated in the presence and absence of MMC (10 nM) are presented. Individual data are shown in dot plots and mean values are represented by bars. Ratios between these numbers indicate the relative MMC resistance of CFCs at different times post-infusion (data shown in Fig. 3). CFC values represent means ± s.e.m. corresponding to the scoring of three plates seeded with BM cells obtained before and after infusion of transduced CD34+ cells. Vector copy numbers per cell obtained in hematopoietic colonies (CVN/CFC) are also represented. An asterisk represents samples not evaluable since the quantity of DNA was under the detection limit for amplification of the human albumin gene by qPCR.
Supplementary information
Supplementary Information
Supplementary Table 1 and Supplementary Fig. 1
Source data
Source Data Fig. 3a and b
Statistical source data Figure 3a and Source Data Figure 3b
Source Data Fig. 3b
Unprocessed immunofluorescence microphotographs
Source Data Extended Data Fig. 2
Statistical Source Data
Source Data Extended Data Fig. 3
Unprocessed microscope images
Source Data Extended Data Fig. 6
Statistical Source Data
Rights and permissions
About this article
Cite this article
Río, P., Navarro, S., Wang, W. et al. Successful engraftment of gene-corrected hematopoietic stem cells in non-conditioned patients with Fanconi anemia. Nat Med 25, 1396–1401 (2019). https://doi.org/10.1038/s41591-019-0550-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-019-0550-z
This article is cited by
-
The transformative potential of HSC gene therapy as a genetic medicine
Gene Therapy (2023)
-
Successes and challenges in clinical gene therapy
Gene Therapy (2023)
-
Hematopoietic reconstitution dynamics of mobilized- and bone marrow-derived human hematopoietic stem cells after gene therapy
Nature Communications (2023)
-
A systematic review and meta-analysis of gene therapy with hematopoietic stem and progenitor cells for monogenic disorders
Nature Communications (2022)
-
Immunological barriers to haematopoietic stem cell gene therapy
Nature Reviews Immunology (2022)