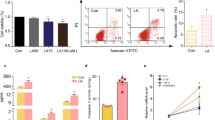
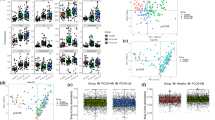
Abstract
Polycystic ovary syndrome (PCOS) is characterized by androgen excess, ovulatory dysfunction and polycystic ovaries1, and is often accompanied by insulin resistance2. The mechanism of ovulatory dysfunction and insulin resistance in PCOS remains elusive, thus limiting the development of therapeutics. Improved metabolic health is associated with a relatively high microbiota gene content and increased microbial diversity3,4. This study aimed to investigate the impact of the gut microbiota and its metabolites on the regulation of PCOS-associated ovarian dysfunction and insulin resistance. Here, we report that Bacteroides vulgatus was markedly elevated in the gut microbiota of individuals with PCOS, accompanied by reduced glycodeoxycholic acid and tauroursodeoxycholic acid levels. Transplantation of fecal microbiota from women with PCOS or B. vulgatus-colonized recipient mice resulted in increased disruption of ovarian functions, insulin resistance, altered bile acid metabolism, reduced interleukin-22 secretion and infertility. Mechanistically, glycodeoxycholic acid induced intestinal group 3 innate lymphoid cell IL-22 secretion through GATA binding protein 3, and IL-22 in turn improved the PCOS phenotype. This finding is consistent with the reduced levels of IL-22 in individuals with PCOS. This study suggests that modifying the gut microbiota, altering bile acid metabolism and/or increasing IL-22 levels may be of value for the treatment of PCOS.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Data relating to the metagenomic sequencing that support the findings of this study have been uploaded to the Sequence Read Archive database (https://trace.ncbi.nlm.nih.gov/Traces/home/) and are available for download via accession number PRJNA530971.
Change history
30 August 2019
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Norman, R. J., Dewailly, D., Legro, R. S. & Hickey, T. E. Polycystic ovary syndrome. Lancet 370, 685–697 (2007).
Dumesic, D. A. et al. Scientific statement on the diagnostic criteria, epidemiology, pathophysiology, and molecular genetics of polycystic ovary syndrome. Endocr. Rev. 36, 487–525 (2015).
Sonnenburg, J. L. & Backhed, F. Diet–microbiota interactions as moderators of human metabolism. Nature 535, 56–64 (2016).
Turnbaugh, P. J., Backhed, F., Fulton, L. & Gordon, J. I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3, 213–223 (2008).
Lindheim, L. et al. Alterations in gut microbiome composition and barrier function are associated with reproductive and metabolic defects in women with polycystic ovary syndrome (PCOS): a pilot study. PLoS ONE 12, e0168390 (2017).
Liu, R. et al. Dysbiosis of gut microbiota associated with clinical parameters in polycystic ovary syndrome. Front. Microbiol. 8, 324 (2017).
Torres, P. J. et al. Gut microbial diversity in women with polycystic ovary syndrome correlates with hyperandrogenism. J. Clin. Endocrinol. Metab. 103, 1502–1511 (2018).
Martin, F. P. et al. A top-down systems biology view of microbiome–mammalian metabolic interactions in a mouse model. Mol. Syst. Biol. 3, 112 (2007).
Claus, S. P. et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol. Syst. Biol. 4, 219 (2008).
Shindo, K., Machida, M., Fukumura, M., Koide, K. & Yamazaki, R. Omeprazole induces altered bile acid metabolism. Gut 42, 266–271 (1998).
Van Houten, E. L. & Visser, J. A. Mouse models to study polycystic ovary syndrome: a possible link between metabolism and ovarian function? Reprod. Biol. 14, 32–43 (2014).
Hand, T. W., Vujkovic-Cvijin, I., Ridaura, V. K. & Belkaid, Y. Linking the microbiota, chronic disease, and the immune system. Trends Endocrinol. Metab. 27, 831–843 (2016).
Tata, B. et al. Elevated prenatal anti-Mullerian hormone reprograms the fetus and induces polycystic ovary syndrome in adulthood. Nat. Med. 24, 834–846 (2018).
Zhong, C. et al. Group 3 innate lymphoid cells continuously require the transcription factor GATA-3 after commitment. Nat. Immunol. 17, 169–178 (2016).
Bouchard, M. F., Taniguchi, H. & Viger, R. S. Protein kinase A-dependent synergism between GATA factors and the nuclear receptor, liver receptor homolog-1, regulates human aromatase (CYP19) PII promoter activity in breast cancer cells. Endocrinology 146, 4905–4916 (2005).
Brestoff, J. R. & Artis, D. Immune regulation of metabolic homeostasis in health and disease. Cell 161, 146–160 (2015).
Bartelt, A. & Heeren, J. Adipose tissue browning and metabolic health. Nat. Rev. Endocrinol. 10, 24–36 (2014).
Yuan, X. et al. Brown adipose tissue transplantation ameliorates polycystic ovary syndrome. Proc. Natl Acad. Sci. USA 113, 2708–2713 (2016).
Zhao, Y. et al. Up-regulated expression of WNT5a increases inflammation and oxidative stress via PI3K/AKT/NF-κB signaling in the granulosa cells of PCOS patients. J. Clin. Endocrinol. Metab. 100, 201–211 (2015).
Dudakov, J. A., Hanash, A. M. & van den Brink, M. R. Interleukin-22: immunobiology and pathology. Annu. Rev. Immunol. 33, 747–785 (2015).
Pedersen, B. K. & Febbraio, M. A. Muscle as an endocrine organ: focus on muscle-derived interleukin-6. Physiol. Rev. 88, 1379–1406 (2008).
Sag, D., Carling, D., Stout, R. D. & Suttles, J. Adenosine 5′-monophosphate-activated protein kinase promotes macrophage polarization to an anti-inflammatory functional phenotype. J. Immunol. 181, 8633–8641 (2008).
Pang, Y. et al. Intermedin restores hyperhomocysteinemia-induced macrophage polarization and improves insulin resistance in mice. J. Biol. Chem. 291, 12336–12345 (2016).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Zhu, W., Lomsadze, A. & Borodovsky, M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 38, e132 (2010).
Perez-Enciso, M. & Tenenhaus, M. Prediction of clinical outcome with microarray data: a partial least squares discriminant analysis (PLS-DA) approach. Hum. Genet. 112, 581–592 (2003).
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2018YFC1003200 and 2018YFC1003900 to Y.P.), National Natural Science Foundation of China (91857115 to C.J.; 81521002 and 81730038 to J.Q.; and 81771650 to R.L.), Clinical Medicine Plus X—Young Scholars Project of Peking University (PKU2019LCXQ025 and PKU2018LCXQ013 to Y.P. and C.J.) and the National Key Research and Development Program of China (2016YFC1000201 to R.L.).
Author information
Authors and Affiliations
Contributions
Y.P., C.J. and J.Q. designed the study and supervised the analyses. X.Q., C.Y., J.X., Y.W., Lina.W., Liying.W., L.M. and Z.Z. enrolled patients and collected patient samples. X.Q., C.Y., L.S., J.X., Q.W., Y. Zhang, X.L. and Y.P. performed the animal experiments. H.L., Y. Zhao, R.L., P.L. and C.Z. helped with the data analyses. F.J.G. and A.D.P. discussed the results. X.Q., C.Y., Y.P., C.J. and J.Q. wrote the manuscript with input from all authors. All authors edited the manuscript and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer Review Information: Randy Levinson and Joao Monteiro were the primary editors on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Gut microbial alterations in women with PCOS.
a,b, Alpha diversity (Shannon index) (a) and beta diversity (Bray–Curtis distance) (b) of the control and PCOS groups at the gene level. For box plots, the midline represents the median, the box represents the interquartile range (IQR) between the first and third quartiles, and whiskers represent the lowest or highest values within 1.5× the IQR from the first or third quartiles. Statistical significance was determined by two-tailed Mann–Whitney U-test. c,d, Top ten bacterial family (c) and bacterial genus (d) abundances in PCOS and control subjects. The q value represents the FDR-adjusted P value. The P value was determined by two-tailed Wilcoxon rank-sum test. In c, for Bacteroidaceae, q = 2.53 × 10−8. In d, for Bacteroides, q = 5.55 × 10−8. Data are presented as means ± s.e.m. Control: n = 43; PCOS: n = 50.
Extended Data Fig. 2 Insulin, hormone, bile acid and IL-22 levels of mice transplanted with PCOS gut microbiota or B. vulgatus.
a, Timeline for the recipient mice transplanted with PCOS or control fecal microbiota. b, Area under the curve (AUC) of GTT (n = 6 mice per group; t10 = −3.032). c, AUC of ITT (n = 6 mice per group; t10 = −2.498). d, Fasting glucose levels (n = 6 mice per group). e, Insulin levels (n = 6 mice per group; t10 = 0.134). f, HOMA-IR (n = 6 mice per group; t10 = −2.669). g, Number of cystic follicles (n = 6 mice per group; t10 = −5.966). h, Number of CLs (n = 6 mice per group; t10 = 7.720; P = 0.000016). i, Levels of testosterone (T) (n = 6 mice per group; t10 = −2.335). j, Levels of luteinizing hormone (n = 6 mice per group; t10 = −5.675). k, Levels of estradiol for the trans-PCOS and trans-control mice (n = 6 mice per group; t10 = 1.322). l, AUC of GTT (n = 8 mice per group; F2,21 = 12.095). m, AUC of ITT (n = 8 mice per group; F2,21 = 5.609). n, Fasting glucose levels (n = 8 mice per group; F2,21 = 6.190). o, Insulin levels (n = 8 mice per group; F2,21 = 4.254). p, HOMA-IR (n = 8 mice per group; F2,21 = 13.559). q, Number of cystic follicles (n = 8 mice per group; F2,21 = 91.000; control versus B. vulgatus, P = 3.17 × 10−9; control versus DHEA, P = 1.74 × 10−11). r, Number of CLs (n = 8 mice per group). s, Levels of T (n = 8 mice per group; control versus DHEA, P = 0.000007). t, Levels of luteinizing hormone (n = 8 mice per group). u, Levels of estradiol for the mice in the control, B. vulgatus gavage or DHEA injection groups (n = 8 mice per group; F2,21 = 18.890; control versus DHEA, P = 0.000039). v, Levels of intestine bile acids (n = 6 mice per group). w, Il22 mRNA expression levels in the intestines, as measured by qPCR (n = 6 mice per group). x, IL-22 levels in the serum of mice transplanted with PCOS or control individual’s fecal microbiota (n = 6 mice per group; t10 = 7.518; P = 0.00002). For d, v and w, P values were determined by two-tailed Mann–Whitney U-test and the data are presented as medians with interquartile ranges. For b, c, e–k and x, P values were determined by two-tailed Student’s t-test and the data are presented as means ± s.e.m. For l–q and u, P values were determined by one-way ANOVA with Tukey’s multiple comparison post-hoc test and data are presented as means ± s.e.m. For r, s and t, P values were determined by Kruskal–Wallis test followed by Dunn’s post-hoc test and data are presented as medians with interquartile ranges.
Extended Data Fig. 3 Improvement of insulin resistance and ovarian dysfunction by administration of IL-22 or bile acid in PCOS mice.
a, Timeline for the mice subjected to IL-22 or GDCA administration after gavage with B. vulgatus. b, Timeline for the PAMH administration PCOS-like mouse model. c, AUC of GTT (n = 6 mice per group; F3,20 = 8.170). d, AUC of ITT (n = 6 mice per group; F3,20 = 10.798; control versus B. vulgatus, P = 0.000035). e, Fasting glucose levels (n = 6 mice per group; F3,20 = 24.524; control versus B. vulgatus, P = 0.000003; B. vulgatus versus B. vulgatus + GDCA, P = 2.18 × 10−7; B. vulgatus versus B. vulgatus + IL-22, P = 0.000002). f, Insulin levels (n = 6 mice per group). g, HOMA-IR (n = 6 mice per group; F3,20 = 48.560; control versus B. vulgatus, P = 1.66 × 10−8; B. vulgatus versus B. vulgatus + GDCA, P = 8.55 × 10−10; B. vulgatus versus B. vulgatus + IL-22, P = 9.47 × 10−9). h, Levels of estradiol for the mice in the control, B. vulgatus, B. vulgatus + GDCA and B. vulgatus + IL-22 groups (n = 6 mice per group; F3,20 = 0.553). i, Levels of estradiol for the mice in the control, PAMH and PAMH + IL-22 groups (n = 6 mice per group; F2,15 = 0.498). For c–e and g–i, P values were determined by one-way ANOVA with Tukey’s multiple comparison post-hoc test and data are presented as means ± s.e.m. For f, the P values were determined by Kruskal–Wallis test followed by Dunn’s post-hoc test and data are presented as medians with interquartile ranges.
Extended Data Fig. 4 Administration of IL-22 prevented insulin resistance and ovarian dysfunction in B. vulgatus-treated mice.
The mice were divided into three groups (control, B. vulgatus and B. vulgatus + IL-22). The animals were gavaged with B. vulgatus or heat-killed B. vulgatus as a control for 3 weeks. At the same time, the mice received IL-22 at a dose of 100 µg kg−1 d−1, intraperitoneally for the B. vulgatus + IL-22 group, or PBS as a parallel control for a–l. a, GTT (n = 8 mice per group). b, AUC of GTT (n = 8 mice per group; F2,21 = 4.076). c, ITT (n = 8 mice per group). d, AUC of ITT (n = 8 mice per group; F2,21 = 23.560; control versus B. vulgatus, P = 0.000036; B. vulgatus versus B. vulgatus + IL-22, P = 0.000002). e, Representative estrous cycles. f, Quantitative analysis of estrous cycles (n = 8 mice per group). g, Number of MII oocytes (n = 8 mice for the control and B. vulgatus + IL-22; n = 10 mice for B. vulgatus; F2,23 = 40.993; control versus B. vulgatus, P = 2.71 × 10−8; B. vulgatus versus B. vulgatus + IL-22, P = 3.40 × 10−7). h, Number of embryos (n = 6 mice per group). i, Immunofluorescence staining for tubulin (green) and DNA (blue) in MII oocytes (MII oocyte number = 150; n = 8 mice for the control and B. vulgatus + IL-22; n = 10 mice for B. vulgatus). Scale bars: 20 μm. Images are representative of two independent experiments with similar results. j, H&E staining of representative ovaries. Scale bars: 200 μm. Images are representative of three independent experiments with similar results. k, Quantitative analysis of cystic follicles (n = 8 mice per group; F2,21 = 37.800; control versus B. vulgatus, P = 2.14 × 10−7; B. vulgatus versus B. vulgatus + IL-22, P = 2.14 × 10−7). l, Quantitative analysis of CLs (n = 8 mice per group; control versus B. vulgatus, P = 0.000045). For a–d, g and k, P values were determined by one-way ANOVA with Tukey’s multiple comparison post-hoc test and data are presented as means ± s.e.m. For f, h and l, P values were determined by the Kruskal–Wallis test followed by Dunn’s post-hoc test and data are presented as medians with interquartile ranges. *P < 0.05; **P < 0.01 versus the control. #P < 0.05; ##P < 0.01 versus B. vulgatus.
Extended Data Fig. 5 Role and mechanisms of bile acid in improving insulin sensitivity and ovarian function.
a, Percentage of IL-22+ cells in CD45+ lineage− RORγT+ ILC3s from siLP of mice with or without TUDCA treatment (n = 4 mice per group; t6 = −5.767). b, mRNA expression of Il22 in the intestines (n = 6 mice per group; t10 = −4.175). c, Serum IL-22 levels of the DHEA or DHEA + GDCA mice (n = 6 mice per group; t10 = −6.067). The mice were divided into three groups (control, GDCA and Il22r−/− + GDCA) for d–i. For the control, wild-type mice were injected daily with DHEA subcutaneously. For GDCA, wild-type mice were injected daily with DHEA subcutaneously and gavaged with 30 mg kg−1 d−1 GDCA. For Il22r−/− + GDCA, Il22r−/− mice were injected daily with DHEA and gavaged with GDCA. d, GTT (n = 6 mice per group). e, AUC of GTT (n = 6 mice per group; F2,15 = 6.293). f, ITT (n = 6 mice per group). g, AUC of ITT (n = 6 mice per group; F2,15 = 14.899). h, Representative estrous cycles for mice from the three groups. i, Quantitative analysis of estrous cycles for the mice from the control, GDCA and Il22r−/− + GDCA groups (n = 6 mice per group). j, IL-22 levels in the cell culture supernatant (n = 4 mice per group; F3,12 = 49.221; DMSO versus GDCA, P = 0.000002; GDCA versus triamterene + GDCA, P = 3.60 × 10−7). k, Il22 mRNA expression levels (n = 4 mice per group; F3,12 = 17.708; GDCA versus triamterene + GDCA, P = 0.000047). l, Gata3 mRNA expression levels in ILC3s with or without GDCA or triamterene treatment in vitro (n = 4 mice per group; F3,12 = 24.044; GDCA versus triamterene + GDCA, P = 0.000012). m, cAMP levels in the intestines of mice treated with DHEA or B. vulgatus (n = 8 mice per group; F2,21 = 29.246; control versus B. vulgatus, P = 3.52 × 10−7; control versus DHEA, P = 0.000013). n, Representative flow cytometry plots of IL-22+ cells in CD45+ lineage− RORγT+ ILC3s from siLP. Three independent experiments were performed with similar results. o, Percentage of IL-22+ cells in CD45+ lineage− RORγT+ ILC3s from siLP of the mice (n = 4 mice per group; F2,9 = 23.175). For a–c, P values were determined by two-tailed Student’s t-test and data are presented as means ± s.e.m. For d–g, j–m and o, P values were determined by one-way ANOVA with Tukey’s multiple comparison post-hoc test and data are presented as means ± s.e.m. For i, P values were determined by Kruskal–Wallis test followed by Dunn’s post-hoc test and data are presented as medians with interquartile ranges.
Extended Data Fig. 6 Adipose tissue browning in PCOS-like mice with or without IL-22 or GDCA treatment.
a–c, Ucp1, Pgc1α, Cited1 and Cox8b mRNA expression levels in brown adipose tissue from: (a) trans-control and trans-PCOS mice (n = 6 mice per group; Ucp1: t10 = 4.789; Pgc1α: t10 = 4.427; Cited1: t10 = 3.846; Cox8b: t10 = 3.153); (b) control, B. vulgatus and DHEA mice (n = 8 mice per group; Ucp1: F2,21 = 14.107; Pgc1α: F2,21 = 8.638, Cited1: F2,21 = 2.612; Cox8b: F2,21 = 9.528); and (c) GDCA or IL-22 therapeutic administration in B. vulgatus-treated mice (control, B. vulgatus, B. vulgatus + GDCA and B. vulgatus + IL-22 mice) (n = 6 mice per group; Ucp1: F3,20 = 19.769, B. vulgatus versus B. vulgatus + IL-22, P = 3.41 × 10−7; Pgc1α: F3,20 = 53.487; control versus B. vulgatus, P = 8.83 × 10−8; B. vulgatus versus B. vulgatus + IL-22, P = 2.39 × 10−10; Cited1: F3,20 = 5.853; Cox8b: F3,20 = 9.242). d,e, Ucp1, Pgc1α, Cited1, Cox8b, Nr2f6 and Prdm16 mRNA expression levels in the subcutaneous fat (d; n = 8 mice per group), and Ucp1, Pgc1α, Cited1 and Cox8b mRNA expression levels in brown adipose tissue (e) after IL-22 preventive administration in B. vulgatus-treated mice (control, B. vulgatus and B. vulgatus + IL-22). For a–e, all of the mice were placed at 4 °C for 12 h before killing. For a, P values were determined by two-tailed Student’s t-test and data are presented as means ± s.e.m. For b and c, P values were determined by one-way ANOVA with Tukey’s multiple comparison post-hoc test and data are presented as means ± s.e.m. For d and e, P values were determined by Kruskal–Wallis test followed by Dunn’s post-hoc test and data are presented as medians with interquartile ranges.
Supplementary information
Supplementary Information
Supplementary Tables 1–3
Source data
Source Data Fig. 1
Statistical Source Data
Source Data Fig. 2
Statistical Source Data
Source Data Fig. 3
Statistical Source Data
Source Data Fig. 4
Statistical Source Data
Source Data Extended Data Fig. 1
Statistical Source Data
Source Data Extended Data Fig. 2
Statistical Source Data
Source Data Extended Data Fig. 3
Statistical Source Data
Source Data Extended Data Fig. 4
Statistical Source Data
Source Data Extended Data Fig. 5
Statistical Source Data
Source Data Extended Data Fig. 6
Statistical Source Data
Rights and permissions
About this article
Cite this article
Qi, X., Yun, C., Sun, L. et al. Gut microbiota–bile acid–interleukin-22 axis orchestrates polycystic ovary syndrome. Nat Med 25, 1225–1233 (2019). https://doi.org/10.1038/s41591-019-0509-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-019-0509-0
This article is cited by
-
Role of polyphenols in remodeling the host gut microbiota in polycystic ovary syndrome
Journal of Ovarian Research (2024)
-
Alterations in gut and genital microbiota associated with gynecological diseases: a systematic review and meta-analysis
Reproductive Biology and Endocrinology (2024)
-
The changing metabolic landscape of bile acids – keys to metabolism and immune regulation
Nature Reviews Gastroenterology & Hepatology (2024)
-
Linoleic acid induces human ovarian granulosa cell inflammation and apoptosis through the ER-FOXO1-ROS-NFκB pathway
Scientific Reports (2024)
-
Future in the past: paternal reprogramming of offspring phenotype and the epigenetic mechanisms
Archives of Toxicology (2024)