Abstract
The gut microbiome is emerging as a key regulator of several metabolic, immune and neuroendocrine pathways1,2. Gut microbiome deregulation has been implicated in major conditions such as obesity, type 2 diabetes, cardiovascular disease, non-alcoholic fatty acid liver disease and cancer3,4,5,6, but its precise role in aging remains to be elucidated. Here, we find that two different mouse models of progeria are characterized by intestinal dysbiosis with alterations that include an increase in the abundance of Proteobacteria and Cyanobacteria, and a decrease in the abundance of Verrucomicrobia. Consistent with these findings, we found that human progeria patients also display intestinal dysbiosis and that long-lived humans (that is, centenarians) exhibit a substantial increase in Verrucomicrobia and a reduction in Proteobacteria. Fecal microbiota transplantation from wild-type mice enhanced healthspan and lifespan in both progeroid mouse models, and transplantation with the verrucomicrobia Akkermansia muciniphila was sufficient to exert beneficial effects. Moreover, metabolomic analysis of ileal content points to the restoration of secondary bile acids as a possible mechanism for the beneficial effects of reestablishing a healthy microbiome. Our results demonstrate that correction of the accelerated aging-associated intestinal dysbiosis is beneficial, suggesting the existence of a link between aging and the gut microbiota that provides a rationale for microbiome-based interventions against age-related diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Sequence data supporting these findings have been deposited in EGA (https://ega-archive.org/) under accession number EGAS00001003656. Metabolomics data are provided in Supplementary Table 5. Any additional data generated and analyzed in this study are available from the corresponding authors upon reasonable request.
References
Thaiss, C. A., Zmora, N., Levy, M. & Elinav, E. The microbiome and innate immunity. Nature 535, 65–74 (2016).
Leulier, F. et al. Integrative physiology: at the crossroads of nutrition, microbiota, animal physiology and human health. Cell Metab. 25, 522–534 (2017).
Koeth, R. A. et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585 (2013).
Loomba, R. et al. Gut microbiome-based metagenomic signature for non-invasive detection of advanced fibrosis in human nonalcoholic fatty liver disease. Cell Metab. 25, 1054–1062 (2017).
Qin, J. et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60 (2012).
Zitvogel, L., Daillere, R., Roberti, M. P., Routy, B. & Kroemer, G. Anticancer effects of the microbiome and its products. Nat. Rev. Microbiol. 15, 465–478 (2017).
Cabreiro, F. & Gems, D. Worms need microbes too: microbiota, health and aging in Caenorhabditis elegans. EMBO Mol. Med. 5, 1300–1310 (2013).
Rios-Covian, D. et al. Intestinal short chain fatty acids and their link with diet and human health. Front. Microbiol. 7, 185 (2016).
Breton, J. et al. Gut commensal E. coli proteins activate host satiety pathways following nutrient-induced bacterial growth. Cell Metab. 23, 324–334 (2016).
Wahlstrom, A., Sayin, S. I., Marschall, H. U. & Backhed, F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 24, 41–50 (2016).
Schroeder, B. O. & Backhed, F. Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 22, 1079–1089 (2016).
Biagi, E. et al. Gut microbiota and extreme longevity. Curr. Biol. 26, 1480–1485 (2016).
O’Toole, P. W. & Jeffery, I. B. Gut microbiota and aging. Science 350, 1214–1215 (2015).
Gordon, L. B., Rothman, F. G., Lopez-Otin, C. & Misteli, T. Progeria: a paradigm for translational medicine. Cell 156, 400–407 (2014).
Puente, X. S. et al. Exome sequencing and functional analysis identifies BANF1 mutation as the cause of a hereditary progeroid syndrome. Am. J. Hum. Genet. 88, 650–656 (2011).
Osorio, F. G. et al. Splicing-directed therapy in a new mouse model of human accelerated aging. Sci. Transl. Med. 3, 106ra107 (2011).
Plovier, H. et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 23, 107–113 (2017).
Schneeberger, M. et al. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci. Rep. 5, 16643 (2015).
Everard, A. et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl Acad. Sci. USA 110, 9066–9071 (2013).
Maharshak, N. et al. Altered enteric microbiota ecology in interleukin 10-deficient mice during development and progression of intestinal inflammation. Gut Microbes 4, 316–324 (2013).
Varela, I. et al. Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature 437, 564–568 (2005).
Rothschild, D. et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215 (2018).
Lloyd-Price, J. et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550, 61–66 (2017).
Goodrich, J. K. et al. Human genetics shape the gut microbiome. Cell 159, 789–799 (2014).
Gopalakrishnan, V. et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018).
Routy, B. et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97 (2018).
Kalabis, J., Rosenberg, I. & Podolsky, D. K. Vangl1 protein acts as a downstream effector of intestinal trefoil factor (ITF)/TFF3 signaling and regulates wound healing of intestinal epithelium. J. Biol. Chem. 281, 6434–6441 (2006).
Barcena, C. et al. Methionine restriction extends lifespan in progeroid mice and alters lipid and bile acid metabolism. Cell Rep. 24, 2392–2403 (2018).
Ridaura, V. K. et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341, 1241214 (2013).
Cox, L. M. et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158, 705–721 (2014).
Vrieze, A. et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 143, 913–916 (2012).
Wang, Y. et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat. Med. 24, 1804–1808 (2018).
van Nood, E. et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 368, 407–415 (2013).
Bodogai, M. et al. Commensal bacteria contribute to insulin resistance in aging by activating innate B1a cells. Sci. Transl. Med. 10, eaat4271 (2018).
Weingarden, A. R. et al. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am. J. Physiol. Gastrointest. Liver Physiol. 306, G310–G319 (2014).
Buffie, C. G. et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 (2015).
Zarrinpar, A. et al. Antibiotic-induced microbiome depletion alters metabolic homeostasis by affecting gut signaling and colonic metabolism. Nat. Commun. 9, 2872 (2018).
de Aguiar Vallim, T. Q., Tarling, E. J. & Edwards, P. A. Pleiotropic roles of bile acids in metabolism. Cell Metab. 17, 657–669 (2013).
Pierre, J. F. et al. Activation of bile acid signaling improves metabolic phenotypes in high-fat diet-induced obese mice. Am. J. Physiol. Gastrointest. Liver Physiol. 311, G286–G304 (2016).
Postler, T. S. & Ghosh, S. Understanding the holobiont: how microbial metabolites affect human health and shape the immune system. Cell Metab. 26, 110–130 (2017).
Wang, C., Li, Q., Redden, D. T., Weindruch, R. & Allison, D. B. Statistical methods for testing effects on ‘maximum lifespan’. Mech. Ageing Dev. 125, 629–632 (2004).
Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011).
Langille, M. G. et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821 (2013).
Abubucker, S. et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput. Biol. 8, e1002358 (2012).
Enot, D. P. et al. Metabolomic analyses reveal that anti-aging metabolites are depleted by palmitate but increased by oleate in vivo. Cell Cycle 14, 2399–2407 (2015).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Osorio, F. G. et al. Nuclear lamina defects cause ATM-dependent NF-κB activation and link accelerated aging to a systemic inflammatory response. Genes Dev. 26, 2311–2324 (2012).
Acknowledgements
The authors thank M. Stamsnijder and the Progeria Family Circle, and all the participants that have kindly engaged in our study. The authors also thank G. Velasco, Y. Español, A.R. Folgueras, X.M. Caravia, J.M. Fraile, I. Varela, M. Mittelbrunn and K. Iribarren for helpful comments and advice; R. Feijoo, A. Moyano, D.A. Puente, S.A. Miranda, M.S. Pitiot, V. García de la Fuente and M.C. Muñiz for excellent technical assistance; M. Gueimonde and C.G. de los Reyes-Gavilán (IPLA-CSIC) for providing A. muciniphila; C. Mayolas for help with logistic organization. The authors acknowledge generous support by J.I. Cabrera. A.M.N. is a recipient of a FPI predoctoral contract and N.S. is supported by a Juan de la Cierva postdoctoral contract, both granted by Ministerio de Economia y Competividad (MINECO). A.L.'s research on aging is funded by Fondo de Investigaciones Sanitarias and Fondos FEDER (PI15/00558). The metabolomics platform is supported by H2020 European Union project OncoBiome. G.K. is supported by Ligue contre le Cancer (équipe labellisée); Agence National de la Recherche (ANR) – Projets blancs; ANR under the frame of E-Rare-2, the ERA-Net for Research on Rare Diseases; Association pour la recherche sur le cancer (ARC); Cancéropôle Ile-de-France; Chancelerie des universités de Paris (Legs Poix), Fondation pour la Recherche Médicale (FRM); a donation by Elior; European Research Area Network on Cardiovascular Diseases (ERA-CVD, MINOTAUR); Gustave Roussy Odyssea; the European Union Horizon 2020 Project Oncobiome; Fondation Carrefour; Institut National du Cancer (INCa); Inserm (HTE); Institut Universitaire de France; LeDucq Foundation; the LabEx Immuno-Oncology; the RHU Torino Lumière; the Seerave Foundation; the SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination (SOCRATE); the SIRIC Cancer Research and Personalized Medicine (CARPEM). The Instituto Universitario de Oncología is supported by Fundación Bancaria Caja de Ahorros de Asturias. J.M.P.F. is supported by Ministerio de Economía y Competitividad-FEDER and Gobierno del Principado de Asturias. C.L.-O. is supported by grants from the European Research Council (DeAge, ERC Advanced Grant), Ministerio de Economía y Competitividad-FEDER, Instituto de Salud Carlos III (RTICC) and Progeria Research Foundation.
Author information
Authors and Affiliations
Contributions
C.B., J.M.P.F., P.M.Q. and C.L.-O. conceived and designed the experiments. R.V.-M., P.M.Q. and C.B. performed bioinformatics analysis and results interpretation. C.B., P.M., C.G., F.R. and P.M.Q. performed experiments and analyzed data. M.T.F.-G. performed histopathological analysis. N.S. and A.M.N. performed the culturing and bacterial solutions of A. muciniphila. N.G. and A.L. organized and carried out pickup of human centenarian samples. S.D., N.B., F.A. and G.K. performed LC-MS-based metabolomics. C.B., J.M.P.F., P.M.Q. and C.L.-O. wrote the manuscript. A.L. and G.K. assisted with manuscript editing and all authors revised the manuscript and provided input.
Corresponding authors
Ethics declarations
Competing interests
G.K. is one of the scientific co-founders of everImmune.
Additional information
Peer review information: Michael Basson and Randy Levinson were the primary editors on this Article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Biodiversity in wild-type (WT) and LmnaG609G/G609G mice at different ages.
a, Scheme representing the microbiome profiling experiment in LmnaG609G/G609G mice. b,c, Comparison of alpha-diversity of gut microbiota among all groups of mice using Shannon’s index (diversity) (b) and the Chao1 index (richness) (c). Analyses were performed using Kruskal–Wallis test (Supplementary Table 1). In the box plots, upper and lower hinges correspond to the first and third quartiles, the center line represents the median, whiskers indicate the highest and lowest values that are within 1.5 × IQR and data beyond the end of the whiskers are outliers and plotted as points. d, Principal coordinates analysis (PCoA) of beta-diversity using the Jaccard distance metric among samples of the five groups analyzed (P < 0.001; PERMANOVA) (Supplementary Table 1). e,f, UPGMA trees representing the beta-diversity analysis of all groups analyzed using the Bray–Curtis dissimilarity (e) and the Jaccard distance metrics (f). For a–e, WT 1mo, n = 6; G609G 1mo, n = 5; WT 4mo, n = 8; G609G 4mo, n = 9; WT 22mo, n = 8.
Extended Data Fig. 2 Statistically significant differences in the gut microbiome of LmnaG609G/G609G versus wild-type (WT) mice at different ages.
a, Total relative abundance of prevalent microbiota at the class level in the five groups studied: 1-month-old WT (n = 6), 1-month-old LmnaG609G/G609G (n = 5), 4-month-old WT (n = 8), 4-month-old LmnaG609G/G609G (n = 9) and 22-month-old WT (n = 8), showing data for individual mice. The low-abundance bacteria group includes all bacterial classes with less than 0.5% of total abundance. b, Results of LEfSe analysis showing bacteria that were significantly different in abundance in progeroid (blue, n = 5) versus WT (violet, n = 6) mice at 1 month of age. c, Results of LEfSe analysis showing bacteria that were significantly different in abundance between WT mice at 1 (violet, n = 6) and 4 (grey, n = 8) months of age. d, Results of LEfSe analysis showing bacteria that were significantly different in abundance between progeroid mice at 1 (blue, n = 5) and 4 (red, n = 9) months of age. e, Results of LEfSe analysis showing bacteria that were significantly different in abundance between WT mice at 4 (grey, n = 8) and 22 (green, n = 8) months of age. f, Results of LEfSe analysis of PICRUSt-HUMAnN2, showing the top five KEGG pathways (based on P value) significantly upregulated in WT and G609G at 4 months (Supplementary Table 3). p, phylum; c, class; o, order; f, family; g, genus. For a–f, analyses were performed with WT 1mo, n = 6; G609G 1mo, n = 5; WT 4mo, n = 8; G609G 4mo, n = 9; WT 22mo, n = 8.
Extended Data Fig. 3 Gut microbiome alteration in progeroid Zmpste24−/− mice.
a, Scheme representing the microbiome profiling experiment in Zmpste24−/− mice. b,c, Comparison of alpha-diversity of gut microbiota between WT and Zmpste24−/− mice using Shannon’s index (diversity) (b) and the Chao1 index (richness) (c). Analyses were performed using the Kruskal–Wallis test (Supplementary Table 1). In the box plots, upper and lower hinges correspond to the first and third quartiles, the center line represents the median, whiskers expand to the highest and lower values that are within 1.5 × IQR and data beyond the end of the whiskers are outliers and plotted as points. d, PCoA of beta-diversity using the Jaccard distance metric between WT and Zmpste24−/− mice (P < 0.002; PERMANOVA). e, Total relative abundance of prevalent microbiota at the class level in Zmpste24−/− and WT mice showing results from individual mice. The low abundance bacteria group includes all bacterial classes with less than 0.5% of total abundance. p, phylum; c, class. For a–e, analyses were performed with WT (n = 8) and Zmpste24−/− (n = 4) mice.
Extended Data Fig. 4 Gut microbiome alteration in human progeria samples.
a, Scheme representing the microbiome profiling experiment in Hutchinson–Gilford (HGPS) and Néstor–Guillermo (NGPS) progeria syndromes. b,c, Comparison of alpha-diversity of gut microbiota using the Shannon’s index (diversity) (b) and the Chao1 index (richness) (c) between progeria patients and healthy siblings. Analyses were performed using the Kruskal–Wallis test (Supplementary Table 1). In the box plots, upper and lower hinges correspond to the first and third quartiles, the center line represents the median, whiskers indicate the highest and lowest values that are within 1.5 × IQR and data beyond the end of the whiskers are outliers and plotted as points. d,e, PCoA of beta-diversity using the Jaccard distances metric among samples analyzed by health status (P = 0.75, PERMANOVA) (d) and by family (P = 0.001, PERMANOVA) (e) (Supplementary Table 1). f, Total relative abundance of prevalent microbiota at the class level in progeria patients and their corresponding healthy relatives, grouped by family and disease condition. The low-abundance bacteria group includes all bacterial classes with less than 0.5% of total abundance. p, phylum; c, class. For a–f, analyses were performed on family A, n = 3 individuals; family B, n = 3 individuals; family C, n = 3 individuals; family D, n = 2 individuals; family E, n = 3 individuals; control (C), n = 9 individuals, progeria (P), n = 5 individuals.
Extended Data Fig. 5 Gut microbiome alteration in human samples from centenarians and healthy controls (adults aged 30–50 years).
a, Scheme representing the microbiome profiling experiment of centenarians and healthy controls. b,c, Comparison of alpha-diversity using Shannon’s index (diversity; P = 0.08) (b) and the Chao1 index (richness; P = 0.00079) (c) in centenarians compared to healthy controls. Analyses were performed with the Kruskal–Wallis test (Supplementary Table 1). In the box plots, upper and lower hinges correspond to the first and third quartiles, the center line represents the median, whiskers indicate the highest and lowest values that are within 1.5 × IQR and data beyond the end of the whiskers are outliers and plotted as points. d, PCoA of beta-diversity using the Jaccard distance metric between centenarians (Ce) and healthy controls (HC) (P < 0.001; PERMANOVA). e, Total relative abundance of prevalent microbiota at the class level in healthy controls (HC) and centenarians (Ce) showing data for each individual. The low-abundance bacteria group includes all bacterial classes with less than 0.5% of total abundance. p, phylum; c, class. For a–e, analyses were performed on centenarians (Ce), n = 17 individuals, and healthy adult controls (HC), n = 14 individuals.
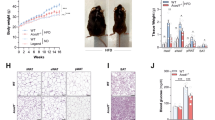
Extended Data Fig. 6 LmnaG609G/G609G mice transplanted with wild-type (WT) gut microbiota show an improved healthspan and lifespan.
a, Average relative abundance of prevalent microbiota at the class level in the FMT experiments, comparing age-matched controls and transplanted mice (n = 8, 4 males and 4 females, in all groups) and donor mice (n = 4, pooled samples from male and female mice). The low-abundance bacteria group includes all bacterial classes representing less than 0.1% of the total. Transplants began at ~6–8 weeks of age and metagenome profiling was analyzed after four rounds of FMT (2 weeks after microbiota depletion). Donor mice, both WT and LmnaG609G/G609G, were ~18–20 weeks old. p, phylum; c, class. b, UPGMA trees representing the beta-diversity analysis of control, transplanted and donor mice in each experimental condition, using Bray–Curtis dissimilarity (Supplementary Table 1). c, Comparison of spleen weight between control WT (n = 4), LmnaG609G/G609G (n = 4), LmnaG609G/G609G mice transplanted with WT microbiota (G609G-WTmic; n = 4) and LmnaG609G/G609G mice transplanted with microbiota from older LmnaG609G/G609G mice (G609G-oG609Gmic n = 4). Analyses were performed with one-way ANOVA with Sidak’s multiple comparison test. Exact adjusted P values are represented within the plot. d, mRNA relative levels (in log10 scale) of inflammatory interleukins in the ileum of WT (n = 9), G609G-control (n = 9), G609G-WTmic (n = 4) and G609G-oG609Gmic (n = 4) mice. Differences were analyzed with ANOVA with Sidak's multiple comparisons test relative to G609G-control mice. Exact adjusted P values are represented within the plot. For c and d, in the box plots, upper and lower hinges correspond to the first and third quartiles, the center line represents the median, whiskers indicate the highest and lowest values that are within 1.5 × IQR, and data beyond the end of the whiskers are outliers and plotted as points. e, Comparison of the number of mice dead and alive between LmnaG609G/G609G (Ctrl; n = 11) and LmnaG609G/G609G mice transplanted with WT microbiota (WTmic; n = 11) at the 80th percentile of the overall survival. Differences were analyzed by one-sized Fisher’s exact test (P = 0.04). f, Percentage survival of LmnaG609G/G609G (n = 11) and LmnaG609G/G609G mice transplanted with buffer (LmnaG609G-EmptyT; n = 8). Transplants started at ~8–10 weeks of age. Differences were analyzed with the log-rank Mantel–Cox test and BH correction was applied after pairwise comparisons between all experimental groups, including those described in Fig. 3f (P = 0.76). Hazard ratio (HR) was calculated using a Cox proportional model (HR of 1.7 [95% confidence interval (CI) 0.64–4.88], P = 0.27). Median and maximal survival are indicated in the Kaplan–Meier plot.
Extended Data Fig. 7 WT mice transplanted with progeroid microbiota show metabolic alterations.
a, Comparison of the body weight between female WT mice transplanted with progeroid microbiota (WT-oG609Gmic; n = 7) and female WT controls (n = 7) over the indicated time period. Differences of body weight over the time were assessed with a linear mixed model and analyzed with an ANOVA type II Wald χ2 test (χ2 = 15.98, d.f. = 1, P = 6.3 × 10−5). Data are presented as mean ± s.e.m. b–d, Metabolic parameters, measured with an Oxymax system, of WT (n = 14; 7 males and 7 females) and WT-oG609Gmic (n = 14; 7 males and 7 females) mice. Comparison of O2 consumption (b), CO2 production (c) and energy expenditure (d) over the indicated period of time. Smoothing curves represent the means of all individuals in each group and shaded areas represent 95% CI.
Extended Data Fig. 8 Zmpste24−/− female mice transplanted with wild-type (WT) gut microbiota show an improved phenotype and extended maximal survival.
a, Comparison of the body weight between control Zmpste24−/− mice (n = 7) and Zmpste24−/− mice transplanted with WT microbiota (Zmpste24−/−-WTmic, n = 7) over the indicated time period. Differences in body weight over the time were assessed with a linear mixed model and analyzed with an ANOVA type II Wald χ2 test (χ2 = 10.52, d.f. = 1, P = 0.0011). b, Comparison of the number of mice dead and alive between Zmpste24−/− (Ctrl; n = 7) and Zmpste24−/− transplanted with WT microbiota (WTmic; n = 7) at the 80th percentile of overall survival. Differences were analyzed by one-sized Fisher’s exact test (P = 0.09).
Supplementary information
Supplementary Information
Supplementary Figures 1–5 and Supplementary Tables 1 and 2
Rights and permissions
About this article
Cite this article
Bárcena, C., Valdés-Mas, R., Mayoral, P. et al. Healthspan and lifespan extension by fecal microbiota transplantation into progeroid mice. Nat Med 25, 1234–1242 (2019). https://doi.org/10.1038/s41591-019-0504-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-019-0504-5
This article is cited by
-
Short-term periodic restricted feeding elicits metabolome-microbiome signatures with sex dimorphic persistence in primate intervention
Nature Communications (2024)
-
The changing metabolic landscape of bile acids – keys to metabolism and immune regulation
Nature Reviews Gastroenterology & Hepatology (2024)
-
Microplastics dampen the self-renewal of hematopoietic stem cells by disrupting the gut microbiota-hypoxanthine-Wnt axis
Cell Discovery (2024)
-
Defective prelamin A processing promotes unconventional necroptosis driven by nuclear RIPK1
Nature Cell Biology (2024)
-
Lactobacillus plantarum A3 attenuates ulcerative colitis by modulating gut microbiota and metabolism
Animal Diseases (2023)