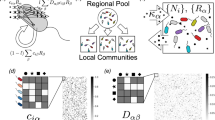
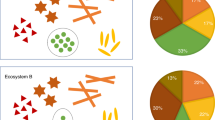
Abstract
The field of human microbiome research has revealed the intimate co-association of humans with diverse communities of microbes in various habitats in the human body, and the necessity of these microbes for the maintenance of human health. Microbial heterogeneity between humans and across spatial and temporal gradients requires multidimensional datasets and a unifying set of theories and statistical tools to analyze the human microbiome and fully realize the potential of this field. Here we consider the utility of community ecology as a framework for the interrogation and interpretation of the human microbiome.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Byrd, A. L. & Segre, J. A. Infectious disease. Adapting Koch’s postulates. Science 351, 224–226 (2016).
Gilbert, J. A. Ecological medicine. Environ. Microbiol. 20, 1917–1919 (2018).
Lederberg, J.M.A. ‘Ome Sweet ’Omics — a genealogical treasury of words. The Scientist (2001).
Gibbons, S. M. et al. Evidence for a persistent microbial seed bank throughout the global ocean. Proc. Natl Acad. Sci. USA 110, 4651–4655 (2013).
Mandal, S. et al. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb. Ecol. Health Dis. 26, 27663 (2015).
Belkaid, Y. & Hand, T. W. Role of the microbiota in immunity and inflammation. Cell 157, 121–141 (2014).
Donaldson, G. P. et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 360, 795–800 (2018).
Donia, M. S. et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158, 1402–1414 (2014).
Garcia-Gutierrez, E., Mayer, M. J., Cotter, P. D. & Narbad, A. Gut microbiota as a source of novel antimicrobials. Gut Microbes 10, 1–21 (2019).
Abisado, R. G., Benomar, S., Klaus, J. R., Dandekar, A. A. & Chandler, J. R. Bacterial quorum sensing and microbial community interactions. MBio 9, e02331–e17 (2018).
Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
Hannigan, G. D., Duhaime, M. B., Koutra, D. & Schloss, P. D. Biogeography and environmental conditions shape bacteriophage-bacteria networks across the human microbiome. PLOS Comput. Biol. 14, e1006099 (2018).
Fujimura, K. E. et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 22, 1187–1191 (2016).
Durack, J. et al. Delayed gut microbiota development in high-risk for asthma infants is temporarily modifiable by Lactobacillus supplementation. Nat. Commun. 9, 707 (2018).
Arrieta, M. C. et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 7, 307ra152 (2015).
Teo, S. M. et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 17, 704–715 (2015).
Fukami, T. & Morin, P. J. Productivity–biodiversity relationships depend on the history of community assembly. Nature 424, 423–426 (2003).
Bäckhed, F. et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 690–703 (2015).
Thaiss, C. A. et al. Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature 540, 544–551 (2016).
Foster, K. R., Schluter, J., Coyte, K. Z. & Rakoff-Nahoum, S. The evolution of the host microbiome as an ecosystem on a leash. Nature 548, 43–51 (2017).
Lynch, S. V. & Pedersen, O. the human intestinal microbiome in health and disease. N. Engl. J. Med. 375, 2369–2379 (2016).
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Rothschild, D. et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215 (2018).
Halfvarson, J. et al. Dynamics of the human gut microbiome in inflammatory bowel disease. New Microbiol. 2, 17004 (2017).
Larsson, P. G. et al. Extended antimicrobial treatment of bacterial vaginosis combined with human lactobacilli to find the best treatment and minimize the risk of relapses. BMC Infect. Dis. 11, 223 (2011).
van Nood, E., Dijkgraaf, M. G. & Keller, J. J. Duodenal infusion of feces for recurrent Clostridium difficile. N. Engl. J. Med. 368, 401–415 (2013).
Kassam, Z., Lee, C. H., Yuan, Y. & Hunt, R. H. Fecal microbiota transplantation for Clostridium difficile infection: systematic review and meta-analysis. Am. J. Gastroenterol. 108, 500–508 (2013).
Panigrahi, P. et al. A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature 548, 407–412 (2017).
Maldonado-Gómez, M. X. et al. Stable engraftment of Bifidobacterium longum AH1206 in the human gut depends on individualized features of the resident microbiome. Cell Host Microbe 20, 515–526 (2016).
Jernberg, C., Löfmark, S., Edlund, C. & Jansson, J. K. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 1, 56–66 (2007).
Suez, J. et al. Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell 174, 1406–1423.e16 (2018).
Zmora, N. et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell 174, 1388–1405.e21 (2018).
Winston, J. A. & Theriot, C. M. Impact of microbial derived secondary bile acids on colonization resistance against Clostridium difficile in the gastrointestinal tract. Anaerobe 41, 44–50 (2016).
Kang, D. W. et al. Microbiota transfer therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome 5, 10 (2017).
Halkjær, S. I. et al. Faecal microbiota transplantation alters gut microbiota in patients with irritable bowel syndrome: results from a randomised, double-blind placebo-controlled study. Gut 67, 2107–2115 (2018).
Cope, E. K., Goldberg, A. N., Pletcher, S. D. & Lynch, S. V. Compositionally and functionally distinct sinus microbiota in chronic rhinosinusitis patients have immunological and clinically divergent consequences. Microbiome 5, 53 (2017).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
Turnbaugh, P. J. et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1, 6ra14 (2009).
Haiser, H. J., Seim, K. L., Balskus, E. P. & Turnbaugh, P. J. Mechanistic insight into digoxin inactivation by Eggerthella lenta augments our understanding of its pharmacokinetics. Gut Microbes 5, 233–238 (2014).
Cussotto, S. et al. Differential effects of psychotropic drugs on microbiome composition and gastrointestinal function. Psychopharmacology (Berl) 1–15 (2018).
Gerber, G. K. The dynamic microbiome. FEBS Lett. 588, 4131–4139 (2014).
Silverman, J., Shenhav, L., Halperin, E., Muklherjee, S. & David, L. Statistical considerations in the design and analysis of longitudinal microbiome studies. Preprint at https://www.biorxiv.org/content/10.1101/448332v1 (2018).
Shenhav, L., Furman, O., Mizrahi, I. & Halperin, E. Modeling the temporal dynamics of the gut microbial community in adults and infants. Preprint at https://doi.org/10.1101/362822 (2017).
Fukuyama, J. et al. Multidomain analyses of a longitudinal human microbiome intestinal cleanout perturbation experiment. PLOS Comput. Biol. 13, e1005706 (2017).
Bucci, V. et al. MDSINE: Microbial Dynamical Systems INference Engine for microbiome time-series analyses. Genome Biol. 17, 121 (2016).
Äijö, T., Müller, C. L. & Bonneau, R. Temporal probabilistic modeling of bacterial compositions derived from 16S rRNA sequencing. Bioinformatics 34, 372–380 (2018).
Silverman, J. D., Durand, H. K., Bloom, R. J., Mukherjee, S. & David, L. A. Dynamic linear models guide design and analysis of microbiota studies within artificial human guts. Microbiome 6, 202 (2018).
Larsen, P. E., Field, D. & Gilbert, J. A. Predicting bacterial community assemblages using an artificial neural network approach. Nat. Methods 9, 621–625 (2012).
Varma, A. P. & Palssom, B. O. Metabolic flux balancing: basic concepts, scientific and practical use. Bio/Technol. 12, 994 (1994).
Yang, L., Yurkovich, J. T., King, Z. A. & Palsson, B. O. Modeling the multi-scale mechanisms of macromolecular resource allocation. Curr. Opin. Microbiol. 45, 8–15 (2018).
Ibarra, R. U., Edwards, J. S. & Palsson, B. O. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature 420, 186–189 (2002).
Zomorrodi, A. R. & Maranas, C. D. OptCom: a multi-level optimization framework for the metabolic modeling and analysis of microbial communities. PLOS Comput. Biol. 8, e1002363 (2012).
Mahadevan, R., Edwards, J. S. & Doyle, F. J. III Dynamic flux balance analysis of diauxic growth in Escherichia coli. Biophys. J. 83, 1331–1340 (2002).
Acknowledgements
The authors would like to thank a number of colleagues for suggestions on articles to include in this review. Specifically, L. P. Coelho; B. Prithiviraj; E. Jasarevic; J. Pickard; M. Yassour; and E. Weinstein. S.V.L. is supported by awards from the National Institutes of Health (AI089473, AI113916, AI133765, PO515267, DH082147, DA040532) and the Crohn’s and Colitis Foundation of America.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
J.A.G. reports roles as Chief Scientific Advisor, 4inno; Scientific Advisory Board, Biome Makers, Inc.; Scientific Advisory Board, DayTwo Inc.; Scientific Advisory Board, Growcentia Inc.; Scientific Advisory Board, Holobiome Inc.; and Scientific Advisory Board, Valent BioSciences. S.V.L. reports research grants from the NIH/NIAID, NIH/NIDA, NIH/NICHD, NIH/Office of the Director and the Crohn’s and Colitis Foundation of America and the following patents and royalties: ‘Reductive prodrug cancer chemotherapy (Stan449-PRV)’; ‘Combination antibiotic and antibody therapy for the treatment of Pseudomonas aeruginosa infection (WO2010091189A1)’ licensed with royalties by KaloBios Inc.; ‘Therapeutic microbial consortium for induction of immune tolerance’ licensed with royalties by Siolta Therapeutics; ‘Systems and methods for detecting antibiotic resistance (WO2012027302A3)’ issued; ‘Nitroreductase enzymes (US7687474B2)’ issued; ‘Sinusitis diagnostics and treatments (WO2013155370A1)’ licensed by Reflourish, LLC; and ‘Methods and systems for phylogenetic analysis (US20120264637A1)’ issued. S.V.L. is a board member of and consultant for Siolta Therapeutics and reports personal fees for this consultancy outside the submitted work.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gilbert, J.A., Lynch, S.V. Community ecology as a framework for human microbiome research. Nat Med 25, 884–889 (2019). https://doi.org/10.1038/s41591-019-0464-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-019-0464-9
This article is cited by
-
A robust microbiome signature for autism spectrum disorder across different studies using machine learning
Scientific Reports (2024)
-
Human gut microbiome, diet, and mental disorders
International Microbiology (2024)
-
Probiotic supplementation during antibiotic treatment is unjustified in maintaining the gut microbiome diversity: a systematic review and meta-analysis
BMC Medicine (2023)
-
Revealing proteome-level functional redundancy in the human gut microbiome using ultra-deep metaproteomics
Nature Communications (2023)
-
The Next Generation of Microbial Ecology and Its Importance in Environmental Sustainability
Microbial Ecology (2023)