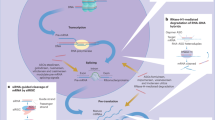
Abstract
Non-alcoholic fatty liver disease ranges from steatosis to non-alcoholic steatohepatitis (NASH), potentially progressing to cirrhosis and hepatocellular carcinoma (HCC). Here, we show that platelet number, platelet activation and platelet aggregation are increased in NASH but not in steatosis or insulin resistance. Antiplatelet therapy (APT; aspirin/clopidogrel, ticagrelor) but not nonsteroidal anti-inflammatory drug (NSAID) treatment with sulindac prevented NASH and subsequent HCC development. Intravital microscopy showed that liver colonization by platelets depended primarily on Kupffer cells at early and late stages of NASH, involving hyaluronan-CD44 binding. APT reduced intrahepatic platelet accumulation and the frequency of platelet–immune cell interaction, thereby limiting hepatic immune cell trafficking. Consequently, intrahepatic cytokine and chemokine release, macrovesicular steatosis and liver damage were attenuated. Platelet cargo, platelet adhesion and platelet activation but not platelet aggregation were identified as pivotal for NASH and subsequent hepatocarcinogenesis. In particular, platelet-derived GPIbα proved critical for development of NASH and subsequent HCC, independent of its reported cognate ligands vWF, P-selectin or Mac-1, offering a potential target against NASH.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Data that support the findings of this study have been uploaded to ArrayExpress (www.ebi.ac.uk/arrayexpress/) and the data set is available under the accession number E-MTAB-6073, entitled ‘Transcriptomic differences in livers of mice fed with normal diet and choline-deficient high-fat diet’.
Change history
18 February 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41591-022-01693-7
References
Fleet, S. E., Lefkowitch, J. H. & Lavine, J. E. Current concepts in pediatric nonalcoholic fatty liver disease. Gastroenterol. Clin. North Am. 46, 217–231 (2017).
European Association for the Study of the Liver, European Association for the Study of Diabetes & European Association for the Study of Obesity. EASL-EASD-EASO Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease. J. Hepatol. 64, 1388–1402 (2016).
Younossi, Z. et al. Global perspectives on non-alcoholic fatty liver disease and non-alcoholic steatohepatitis. Hepatology https://doi.org/10.1002/hep.30251 (2018).
Brunt, E. M. et al. Nonalcoholic fatty liver disease. Nat. Rev. Dis. Primers 1, 15080 (2015).
Friedman, S. L., Neuschwander-Tetri, B. A., Rinella, M. & Sanyal, A. J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 24, 908–922 (2018).
El-Serag, H. B. & Kanwal, F. Epidemiology of hepatocellular carcinoma in the United States: where are we? Where do we go? Hepatology 60, 1767–1775 (2014).
Torre, L. A. et al. Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108 (2015).
Ozakyol, A. Global epidemiology of hepatocellular carcinoma (HCC epidemiology). J. Gastrointest. Cancer 48, 238–240 (2017).
Ashfin, A. et al. Health effects of overweight and obesity in 195 countries over 25 years. N. Engl. J. Med. 377, 13–27 (2017).
Ringelhan, M., Pfister, D., O’Connor, T., Pikarsky, E. & Heikenwalder, M. The immunology of hepatocellular carcinoma. Nat. Immunol. 19, 222–232 (2018).
Wolf, M. J. et al. Metabolic activation of intrahepatic CD8+ T cells and NKT cells causes nonalcoholic steatohepatitis and liver cancer via cross-talk with hepatocytes. Cancer Cell. 26, 549–564 (2014).
Michelson, A. D. How platelets work: platelet function and dysfunction. J. Thromb. Thrombolysis 16, 7–12 (2003).
Chauhan, A., Adams, D. H., Watson, S. P. & Lalor, P. F. Platelets: no longer bystanders in liver disease. Hepatology 64, 1774–1784 (2016).
Gawaz, M., Langer, H. & May, A. E. Platelets in inflammation and atherogenesis. J. Clin. Invest. 115, 3378–3384 (2005).
Jackson, S. P. Arterial thrombosis—insidious, unpredictable and deadly. Nat. Med. 17, 1423–1436 (2011).
Fujita, K. et al. Effectiveness of antiplatelet drugs against experimental non-alcoholic fatty liver disease. Gut 57, 1583–1591 (2008).
Lang, P. A. et al. Aggravation of viral hepatitis by platelet-derived serotonin. Nat. Med. 14, 756–761 (2008).
Iannacone, M. et al. Platelets mediate cytotoxic T lymphocyte-induced liver damage. Nat. Med. 11, 1167–1169 (2005).
Iannacone, M., Sitia, G., Narvaiza, I., Ruggeri, Z. M. & Guidotti, L. G. Antiplatelet drug therapy moderates immune-mediated liver disease and inhibits viral clearance in mice infected with a replication-deficient adenovirus. Clin. Vaccine Immunol. 14, 1532–1535 (2007).
Sitia, G. et al. Antiplatelet therapy prevents hepatocellular carcinoma and improves survival in a mouse model of chronic hepatitis B. Proc. Natl Acad. Sci. USA 109, E2165–E2172 (2012).
Shen, H., Shahzad, G., Jawairia, M., Bostick, R. M. & Mustacchia, P. Association between aspirin use and the prevalence of nonalcoholic fatty liver disease: a cross-sectional study from the third national health and nutrition examination survey. Aliment. Pharmacol. Ther. 40, 1066–1073 (2014).
Musso, G., Cassader, M., Rosina, F. & Gambino, R. Impact of current treatments on liver disease, glucose metabolism and cardiovascular risk in non-alcoholic fatty liver disease (NAFLD): a systematic review and meta-analysis of randomised trials. Diabetologia 55, 885–904 (2012).
Wong, V. W. et al. Pathogenesis and novel treatment options for non-alcoholic steatohepatitis. Lancet Gastroenterol. Hepatol. 1, 56–67 (2016).
Clapper, J. R. et al. Diet-induced mouse model of fatty liver disease and nonalcoholic steatohepatitis reflecting clinical disease progression and methods of assessment. Am. J. Physiol. Gastrointest. Liver Physiol. 305, G483–G495 (2013).
Wang, B. et al. Intestinal phospholipid remodeling is required for dietary-lipid uptake and survival on a high-fat diet. Cell. Metab. 23, 492–504 (2016).
Weston, C. J. et al. Vascular adhesion protein-1 promotes liver inflammation and drives hepatic fibrosis. J. Clin. Invest. 125, 501–520 (2015).
Gomes, A. L. et al. Metabolic inflammation-associated il-17a causes non-alcoholic steatohepatitis and hepatocellular carcinoma. Cancer Cell. 30, 161–175 (2016).
Angulo, P. et al. Liver fibrosis, but no other histologic features, is associated with long-term outcomes of patients with nonalcoholic fatty liver disease. Gastroenterology 149, 389–397 e310 (2015).
Dulai, P. S. et al. Increased risk of mortality by fibrosis stage in nonalcoholic fatty liver disease: Systematic review and meta-analysis. Hepatology 65, 1557–1565 (2017).
Bayindir-Buchhalter, I. et al. Cited4 is a sex-biased mediator of the antidiabetic glitazone response in adipocyte progenitors. EMBO Mol. Med. 10, e8613 (2018).
Mauri, L. et al. Twelve or 30 months of dual antiplatelet therapy after drug-eluting stents. N. Engl. J. Med. 371, 2155–2166 (2014).
He, G. & Karin, M. NF-kappaB and STAT3—key players in liver inflammation and cancer. Cell Res. 21, 159–168 (2011).
Kral, J. B., Schrottmaier, W. C., Salzmann, M. & Assinger, A. Platelet interaction with innate immune cells. Transfus. Med. Hemother. 43, 78–88 (2016).
Husted, S. & van Giezen, J. J. Ticagrelor: the first reversibly binding oral P2Y12 receptor antagonist. Cardiovasc. Ther. 27, 259–274 (2009).
Wallentin, L. et al. Ticagrelor versus clopidogrel in patients with acute coronary syndromes. N. Engl. J. Med. 361, 1045–1057 (2009).
Deppermann, C. et al. Gray platelet syndrome and defective thrombo-inflammation in Nbeal2-deficient mice. J. Clin. Invest. 123, 3331–3342 (2013).
Albers, C. A. et al. Exome sequencing identifies NBEAL2 as the causative gene for gray platelet syndrome. Nat. Genet. 43, 735–737 (2011).
Kahr, W. H. et al. Mutations in NBEAL2, encoding a BEACH protein, cause gray platelet syndrome. Nat. Genet. 43, 738–740 (2011).
Gunay-Aygun, M. et al. NBEAL2 is mutated in gray platelet syndrome and is required for biogenesis of platelet alpha-granules. Nat. Genet. 43, 732–734 (2011).
Kopec, A. K. et al. Thrombin promotes diet-induced obesity through fibrin-driven inflammation. J. Clin. Invest. 127, 3152–3166 (2017).
Jandrot-Perrus, M. et al. Cloning, characterization, and functional studies of human and mouse glycoprotein VI: a platelet-specific collagen receptor from the immunoglobulin superfamily. Blood 96, 1798–1807 (2000).
Drescher, H. K. et al. beta7-Integrin and MAdCAM-1 play opposing roles during the development of non-alcoholic steatohepatitis. J. Hepatol. 66, 1251–1264 (2017).
Haemmerle, M., Stone, R. L., Menter, D. G., Afshar-Kharghan, V. & Sood, A. K. The platelet lifeline to cancer: challenges and opportunities. Cancer Cell. 33, 965–983 (2018).
Kleinschnitz, C. et al. Targeting platelets in acute experimental stroke: impact of glycoprotein Ib, VI, and IIb/IIIa blockade on infarct size, functional outcome, and intracranial bleeding. Circulation 115, 2323–2330 (2007).
Kanaji, T., Russell, S. & Ware, J. Amelioration of the macrothrombocytopenia associated with the murine Bernard-Soulier syndrome. Blood 100, 2102–2107 (2002).
Subramaniam, M. et al. Defects in hemostasis in P-selectin-deficient mice. Blood 87, 1238–1242 (1996).
Blenner, M. A., Dong, X. & Springer, T. A. Structural basis of regulation of von Willebrand factor binding to glycoprotein Ib. J. Biol. Chem. 289, 5565–5579 (2014).
Wang, Y. et al. Leukocyte integrin Mac-1 regulates thrombosis via interaction with platelet GPIbalpha. Nat. Commun. 8, 15559 (2017).
George, J. N. Platelets. Lancet 355, 1531–1539 (2000).
Langer, H. F. et al. Platelets contribute to the pathogenesis of experimental autoimmune encephalomyelitis. Circ. Res. 110, 1202–1210 (2012).
Romo, G. M. et al. The glycoprotein Ib-IX-V complex is a platelet counterreceptor for P-selectin. J. Exp. Med. 190, 803–814 (1999).
Kroll, M. H., Harris, T. S., Moake, J. L., Handin, R. I. & Schafer, A. I. von Willebrand factor binding to platelet GpIb initiates signals for platelet activation. J. Clin. Invest. 88, 1568–1573 (1991).
Wong, J. et al. A minimal role for selectins in the recruitment of leukocytes into the inflamed liver microvasculature. J. Clin. Invest. 99, 2782–2790 (1997).
Dutting, S. et al. A Cdc42/RhoA regulatory circuit downstream of glycoprotein Ib guides transendothelial platelet biogenesis. Nat. Commun. 8, 15838 (2017).
Min, H. K. et al. Increased hepatic synthesis and dysregulation of cholesterol metabolism is associated with the severity of nonalcoholic fatty liver disease. Cell Metab. 15, 665–674 (2012).
Townsend, S. A. & Newsome, P. N. Non-alcoholic fatty liver disease in 2016. Br. Med. Bull. 119, 143–156 (2016).
Neuschwander-Tetri, B. A. et al. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet 385, 956–965 (2015).
Boege, Y. et al. A dual role of caspase-8 in triggering and sensing proliferation-associated dna damage, a key determinant of liver cancer development. Cancer Cell. 32, 342–359 e310 (2017).
Franchi, F., Rollini, F. & Angiolillo, D. J. Antithrombotic therapy for patients with STEMI undergoing primary PCI. Nat. Rev. Cardiol. 14, 361–379 (2017).
Margetts, J. et al. Neutrophils: driving progression and poor prognosis in hepatocellular carcinoma? Br. J. Cancer 118, 248–257 (2018).
Bender, M., Hagedorn, I. & Nieswandt, B. Genetic and antibody-induced glycoprotein VI deficiency equally protects mice from mechanically and FeCl3-induced thrombosis. J. Thromb. Haemost. 9, 1423–1426 (2011).
Tiedt, R., Schomber, T., Hao-Shen, H. & Skoda, R. C. Pf4-Cre transgenic mice allow the generation of lineage-restricted gene knockouts for studying megakaryocyte and platelet function in vivo. Blood 109, 1503–1506 (2007).
World Medical Association Declaration of Helsinki. Recommendations guiding physicians in biomedical research involving human subjects. Cardiovasc. Res. 35, 2–3 (1997).
International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use. ICH harmonized tripartite guideline: guideline for good clinical practice. J. Postgrad. Med. 47, 45–50 (2001).
European Commission. Directive 2001/20/EC of the European Parliament and of the Council of 4 April 2001 on the approximation of the laws, regulations and administrative provisions of the member states relating to the implementation of good clinical practice in the conduct of clinical trials on medicinal products for human use. Med Etika Bioet. 9, 12–19 (2002).
Grundy, S. M. et al. Implications of recent clinical trials for the national cholesterol education program adult treatment panel III guidelines. Circulation 110, 227–239 (2004).
Werner, M. et al. All-In-One: advanced preparation of human parenchymal and non-parenchymal liver cells. PLoS One 10, e0138655 (2015).
Maaten, L. J. Pvd & Hinton, G. E. Visualizing high-dimensional data using t-SNE. J Mach Learn Res. 9, 2579–2605 (2008).
Raasch, J. et al. IkappaB kinase 2 determines oligodendrocyte loss by non-cell-autonomous activation of NF-kappaB in the central nervous system. Brain 134, 1184–1198 (2011).
Bergmeier, W. et al. Flow cytometric detection of activated mouse integrin alphaIIbbeta3 with a novel monoclonal antibody. Cytometry 48, 80–86 (2002).
Schulte, V. et al. Targeting of the collagen-binding site on glycoprotein VI is not essential for in vivo depletion of the receptor. Blood 101, 3948–3952 (2003).
Nieswandt, B. et al. Long-term antithrombotic protection by in vivo depletion of platelet glycoprotein VI in mice. J. Exp. Med. 193, 459–469 (2001).
Gruner, S. et al. Multiple integrin-ligand interactions synergize in shear-resistant platelet adhesion at sites of arterial injury in vivo. Blood 102, 4021–4027 (2003).
Nieswandt, B., Bergmeier, W., Rackebrandt, K., Gessner, J. E. & Zirngibl, H. Identification of critical antigen-specific mechanisms in the development of immune thrombocytopenic purpura in mice. Blood 96, 2520–2527 (2000).
May, F. et al. CLEC-2 is an essential platelet-activating receptor in hemostasis and thrombosis. Blood 114, 3464–3472 (2009).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Parekh, S., Ziegenhain, C., Vieth, B., Enard, W. & Hellmann, I. The impact of amplification on differential expression analyses by RNA-seq. Sci. Rep. 6, 25533 (2016).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Levin, Y. S. et al. Effect of echo-sampling strategy on the accuracy of out-of-phase and in-phase multiecho gradient-echo MRI hepatic fat fraction estimation. J. Magn. Reson. Imaging 39, 567–575 (2014).
Merkle, E. M. & Nelson, R. C. Dual gradient-echo in-phase and opposed-phase hepatic MR imaging: a useful tool for evaluating more than fatty infiltration or fatty sparing. Radiographics 26, 1409–1418 (2006).
Nombela-Arrieta, C. et al. Quantitative imaging of haematopoietic stem and progenitor cell localization and hypoxic status in the bone marrow microenvironment. Nat. Cell Biol. 15, 533–543 (2013).
Thon, J. N. & Italiano, J. E. Platelets: production, morphology and ultrastructure. Handb. Exp. Pharmacol. 210, 3–22 (2012).
Thon, J. N. & Italiano, J. E. Jr. Does size matter in platelet production? Blood 120, 1552–1561 (2012).
Acknowledgements
We thank D. Heide, J. Hetzer, R. Hillermann, C. Gropp, F. Müller, S. Prokosch, D. Kull, R. Dunkl, O. Seelbach, M. Bawohl, R. Maire, M. Bieri, C. Mittmann, H. Honcharova-Biletska, A. Fitsche, A. Adili, P. Münzer, T. Nussbaumer, F. Prutek, G. Dharmalingam and I. Singh for excellent technical assistance. We thank K. Nikolaou for the help with the human cohort recruitment and analysis. M. Malehmir was partially supported by grants from the University Zurich (Zurich Integrative Human Physiology (ZHIP) Sprint Fellowship) and from the Hartmann Müller Stiftung, Zurich. A.W. was supported by a grant from the Swiss National Science Foundation (320030_182764/1). M. Heikenwaelder was supported by an ERC Consolidator grant (HepatoMetaboPath), an EOS grant, SFBTR 209, SFBTR179, Research Foundation Flanders (FWO) under grant 30826052 (EOS Convention MODEL-IDI), Deutsche Krebshilfe projects 70113166 and 70113167, and the Helmholtz-Gemeinschaft, Zukunftsthema ‘Immunology and Inflammation’ (ZT-0027). This project has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement 667273 and the DFG (SFB/TR 240 (project 374031971) to B.N. and D.S.), ERC Consolidator grant ‘CholangioConcept’ (to L.Z.), and the German Research Foundation (DFG): grants FOR2314, SFB685 and the Gottfried Wilhelm Leibniz Program (to L.Z.). Further funding was provided by the German Ministry for Education and Research (BMBF) (eMed/Multiscale HCC), the German Universities Excellence Initiative (third funding line: ‘future concept’), the German Center for Translational Cancer Research (DKTK) and the German-Israeli Cooperation in Cancer Research (DKFZ-MOST) (to L.Z. and M. Heikenwaelder). D.I. was supported by an EMBO Long-term Fellowship. J.M.L. is supported by Asociación Española Contra el Cáncer (Accelerator award: HUNTER), Spanish National Health Institute (SAF2013–41027), Generalitat de Catalunya (SGR 1162 and AGAUR, SGR-1358), the Samuel Waxman Cancer Research Foundation, the US Department of Defense (CA150272P3), the European Commission Horizon 2020 Program (HEPCAR, proposal number 667273-2), and the National Cancer Institute (P30 CA196521). D.A.M. is supported by CRUK grant C18342/A23390 and MRC grant MR/K001949/1. M.P. is supported by the German Research Foundation (DFG). M.G., T.G. and D.R. was supported by grants from the German Research Foundation (KFO274 and SFB/TR240 (project 374031971)). D.J.W. received a Wellcome Trust Strategic Award (098565/Z/12/Z) and funding from the Medical Research Council (MC-A654-5QB40). C.L.W. was funded by CRUK project Cancer Research UK Programme Grant C18342/A23390. H.G.A. has been supported by the Deutsche Forschungsgemeinschaft (SFB-TR209 ‘Liver Cancer’).
Author information
Authors and Affiliations
Contributions
M. Malehmir, D.P., S.G., M.S. and D.I. contributed equally as first authors. E.K., V.L., M.P. and B.G.J.S. contributed equally as second authors. Design of the study: M. Malehmir, M.J.W., D.R., A.W., B.N., M.G. and M. Heikenwaelder. M. Malehmir, E.K., D.P., V.L., M.J.W. and C.D. performed breeding and housing of mice. M. Malehmir, S.G., M.S., E.K., D.P., V.L., D.I., A.A., M.P., B.G.J.S., A.O., C.D., J.V., D.S., D.D., C.L.W., P.H., A.R., A.T., H.D., O.K., M.K., C.J.W., A.C., R.B., N.A., M.E.H., L.S. and M. Hinterleitner performed experiments. D.R., M.R., F.B., T.G., M.N.B., O.B., M.N. and M.G. designed and performed the clinical case study. J.W., R.P., N.D., L.Z., D.J.W, H.G.A, H.D., D.K., F.T., P.F.L., T.O., D.J.W., A.V., M.D.M., A.J.R., R.R., P.K., P.A.K., B.N., A.W., J.M.L, M. Matter, D.A.M., T.S., M.P., L.S., D.H.A., C.N.-A. and J.L. provided tissue samples or mouse strains and/or scientific input. K.U. and T.E. performed biostatistical analyses. All authors analyzed data. M. Malehmir, M.E.H., D.P., S.G., M.S., P.K., B.N., M.G., O.K., T.O., A.W., and M. Heikenwaelder wrote the manuscript, and all authors contributed to writing and provided feedback.
Corresponding authors
Ethics declarations
Competing interests
J.M.L. receives consulting fees from Bayer HealthCare Pharmaceuticals, Eli Lilly, Bristol-Myers Squibb, Merck, Eisai Inc, Celsion Corporation, Exelixis, Merck, Ipsen, Glycotest, Navigant, Leerink Swann LLC, Midatech Ltd, Fortress Biotech, Sprink Pharmaceuticals and Nucleix and research support from Bayer HealthCare Pharmaceuticals, Eisai Inc, Bristol-Myers Squibb and Ipsen. This article presents independent research supported in part by the National Institute for Health Research (NIHR) Birmingham Biomedical Research Centre. The views expressed are those of the author(s) and not necessarily those of the National Health Service, the NIHR, or the Department of Health.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Transcriptional alterations in the course of NASH and physical interaction of immune cells, liver endothelium and platelets.
a. GSEA and expression analysis of genes related clusters of platelet activation, aggregation, degranulation and genes associated to TNFα family regulation, cytokine interaction and lymphocyte migration from 6 months ND-fed versus CD-HFD-fed mice. b–g, Gene expression alterations found in annotated genes of indicated clusters and mice shown in a. h, Gene expression alterations found in annotated genes of indicated cluster of cell chemotaxis (left) and GSEA and expression analysis of genes associated to the cell chemotaxis cluster from mice shown in a (right). i, Transcriptional alterations for genes associated with platelet activation/aggregation of the 7.5 month ND versus WD-HTF fed mice.
Extended Data Fig. 2 Sulindac treatment does not prevent NASH pathology.
a, Left, mouse weight development in 12 months ND-, CD-HFD- or CD-HFD/sulindac-fed mice (n = 5 mice/group). Statistics: ND vs CD-HFD (black asterisks), ND vs CD-HFD/sulindac (red asterisks). Middle, liver/body weight (6 months: n = 5 mice/group; 12 months: ND n = 3 mice; CD-HFD n = 8 mice; CD-HFD/sulindac n = 7 mice) and (right) ALT levels of 6 and 12 months ND-, CD-HFD- or CD-HFD/sulindac-fed mice (6 months: ND n = 3 mice; CD-HFD n = 4 mice; CD-HFD/sulindac n = 5 mice; 12 months: ND n = 7 mice; CD-HFD n = 12 mice; CD-HFD/sulindac = 5 mice). b, (left) Liver triglycerides and (middle) serum cholesterol levels in 6 and 12 months ND-, CD-HFD- or CD-HFD/sulindac-fed mice (liver TGs 6 months: ND = 7 mice; CD-HFD n = 11 mice; CD-HFD/sulindac n = 4 mice; Liver TGs 12 months: ND n= 4 mice; CD-HFD n = 9 mice; CD-HFD/sulindac n = 10 mice; cholesterol 6 months: ND n = 4 mice; CD-HFD n = 4 mice; CD-HFD/sulindac n = 5 mice; cholesterol 12 months: ND n = 6 mice; CD-HFD n = 10 mice; CD-HFD/sulindac n = 10 mice). (right) IPGTT performed with 6 months ND-, CD-HFD- or CD-HFD/sulindac-fed mice (n = 5 mice/group). Statistics: ND vs CD-HFD (black asterisks). (c) MRI analyses of livers of 6 months ND-, CD-HFD- or CD-HFD/sulindac-fed mice (n = 3 mice/group). d, (left) Analysis by H&E indicate damaged hepatocytes (asterisk) and (right) evaluation by NAS in livers of 6 months CD-HFD- or CD-HFD/sulindac-fed mice (ND n = 9 mice; CD-HFD n = 9 mice; CD-HFD/sulindac n = 10 mice), scale bars: 100 µm in 10×, 50 µm in 20×. e, Real-time qPCR analysis for mRNA of genes involved in lipid metabolism in liver of 6 months ND-, CD-HFD- or CD-HFD/sulindac-fed mice (ND n = 4 mice; CD-HFD n = 5 mice; CD-HFD/sulindac n = 3 mice). Comparison between CD-HFD vs CD-HFD/sulindac. f, Sudan red staining and quantification for fat accumulation of 6 months ND-, CD-HFD- or CD-HFD/sulindac-fed mice (ND n = 5; CD-HFD n = 4; CD-HFD/sulindac n = 5). Scale bar: 100 µm. All data are shown as mean ± s.e.m. Data in a (left) and b (right) were analyzed by two-way analysis of variance with post hoc Tukey’s multiple comparison test. Data in a (middle and right), b (left and middle), d and f were analyzed by one-way analysis of variance with post hoc Tukey’s multiple comparison test. Data in e were analyzed by two-tailed Mann-Whitney t test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Fig. 3 Ticagrelor treatment attenuates CD-HFD-induced NASH and NASH-associated conditions, and prevents HCC.
a, CD42b staining and quantification in 6 months CD-HFD or CD-HFD/Ticagrelor fed mice (CD-HFD n = 4 mice; CD-HFD/Ticagrelor n = 8 mice), scale bar: 50 µm. b, 3D confocal images of platelet (green)/liver endothelium (grey) interaction in livers of 6 months ND, CD-HFD and CD-HFD/Ticagrelor fed mice (n = 4 mice/group), scale bar: 20 μm. Quantification of platelet (PLT) aggregate size, overall PLT surface and quantification of platelet/liver endothelium coverage in focus of view (n = 4 mice/group). For visualization of intravascular events, the transparency of the sinusoidal rendering was set to 50%. c, Body weight development in 12 months ND, CD-HFD or CD-HFD/Ticagrelor fed mice (ND n = 4 mice; CD-HFD n = 6 mice; CD-HFD/Ticagrelor n = 4 mice). Statistics: ND vs CD-HFD (black asterisks), ND vs CD-HFD/Ticagrelor (green asterisks). d–h ALT (d), Liver triglyceride (e), serum cholesterol levels (f), VLDL secretion in serum (g) and IPGTT (h) of 6 months ND, CD-HFD or CD-HFD/Ticagrelor fed mice (ALT: ND n = 8 mice; CD-HFD n = 5 mice; CD-HFD/Ticagrelor n = 10 mice; liver triglycerides: ND n = 7 mice; CD-HFD n = 11 mice; CD-HFD/Ticagrelor n = 6 mice; serum cholesterol: n = 8 mice/group; liver triglycerides: ND n = 8 mice; CD-HFD n = 11 mice; CD-HFD/Ticagrelor n = 8 mice; IPGTT: ND n = 5 mice; CD-HFD n = 3 mice; CD-HFD/Ticagrelor n = 3 mice). i,j, Real-time qPCR analysis of hepatic genes associated to catabolic (i) and anabolic processes (j) of lipid metabolism of 6 months ND, CD-HFD or CD-HFD/Ticagrelor fed mice (RT-qPCR for catabolic genes: ND n = 2 mice; CD-HFD n = 6 mice; CD-HFD/Ticagrelor n = 6 mice; RT-qPCR for anabolic genes: ND n = 2 mice; CD-HFD n = 3 mice; CD-HFD/Ticagrelor n = 3 mice). All data are shown as mean ± s.e.m. Data in a were analyzed by two-tailed Student's t test. Data in b, d, e, f, g and j were analyzed by one way ANOVA with the post hoc Tukey’s multiple comparison test. Data in c and h were analyzed by two way ANOVA with the post hoc Tukey’s multiple comparison test. Data in i were analyzed by two-tailed Mann–Whitney test, N.s.: not significant *P < 0.05. **P < 0.01. ***P < 0.001. ****P < 0.0001.
Extended Data Fig. 4 Mice with non-functional platelet aggregation are not protected from NASH development.
a, CD42b staining and quantification of 6 months CD-HFD- or CD-HFD/Itg2b−/−-fed mice (CD-HFD n = 5 mice, CD-HFD/Itg2b–/–n = 4 mice). b, Body weight development of 6 months CD-HFD or CD-HFD/Itg2b−/−-fed mice (n = 5/group). Statistics: ND vs CD-HFD (black asterisks), ND vs CD-HFD/Itg2b–/– (blue asterisks). c, ALT (ND n = 4 mice, CD-HFD n = 3 mice, CD-HFD/Itg2b–/–n = 3 mice), AST (ND n = 5 mice, CD-HFD n = 3 mice, CD-HFD/Itg2b–/–n = 3 mice), (d) liver triglycerides (ND n = 7 mice, CD-HFD n = 8 mice, CD-HFD/Itg2b–/–n = 3 mice), and serum cholesterol levels (ND n = 7 mice, CD-HFD n= 3 mice, CD-HFD/Itg2b–/–n = 3 mice), (e) IPGTT from mice shown in a (n = 3–4 field/mouse, ND n = 4 mice, CD-HFD n= 4 mice, CD-HFD/Itg2b–/–n = 10 mice) Statistics: ND vs CD-HFD (black asterisks), ND vs CD-HFD/Itg2b–/– (blue asterisks). (f) Real-time qPCR analysis for genes involved in lipid metabolism/β-oxidation (ND n = 2 mice, CD-HFD n = 4 mice, CD-HFD/Itg2b–/–n = 3 mice). Statistics: CD-HFD vs CD-HFD/tg2b–/– (blue asterisks). (g) NAS evaluation (CD-HFD n = 9 mice, CD-HFD/tg2b–/–n = 7 mice) and (h) quantification of fat by Sudan red staining of mice shown in a (n = 5 mice/group), scale bar: 100 µm in 10×, 50 µm in 20×. All data are shown as mean ± s.e.m. Data shown in a and g were analyzed by two-tailed Student’s t test. Data in b,e were analyzed by two-way analysis of variance with the post hoc Bonferroni multiple comparison test. Data in c,d,h were analyzed by one-way analysis of variance with the post hoc Tukey’s multiple comparison test. Data in f were analyzed by two-tailed Mann-Whitney test; n.s., not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Fig. 5 Gp6 mice display severe steatosis, NASH and NASH-associated conditions.
a, CD42b staining and quantification of 6 months CD-HFD or CD-HFD/Gp6–/–-fed mice (CD-HFD n = 5 mice, CD-HFD/Gp6-–/–n = 4 mice), scale bar: 50 µm. b, Body weight (n = 6 mice/group), (c) ALT, liver triglycerides and cholesterol levels of 6 months mice (ND n = 4 mice, CD-HFD n = 4 mice, CD-HFD/Gp6–/–n = 3 mice). Statistics: ND vs CD-HFD (black asterisks), ND vs CD-HFD/Gp6 (orange asterisks). d, IPGTT (ND n = 5 mice, CD-HFD n = 5 mice, CD-HFD/Gp6–/–n = 3 mice). Statistics: ND vs CD-HFD (black asterisks), ND vs CD-HFD/Gp6–/– (orange asterisks). e, Real-time qPCR analysis for genes involved in lipid metabolism/β-oxidation (ND n = 2 mice, CD-HFD n = 4 mice, CD-HFD/Gp6–/–n = 3 mice). f, NAS evaluation of mice shown in a, damaged hepatocytes are indicated by asterisks (CD-HFD n = 9 mice, CD-HFD/Gp6–/– n = 8 mice), scale bar: 100 µm in 10× and 50 µm in 20×. g, Quantification of fat by Sudan red staining in mice shown in b (n = 3–4 fields/mouse, ND n = 4 mice, CD-HFD n = 4 mice, CD-HFD/Gp6–/–n = 9 mice). All data are shown as mean ± s.e.m. Data in b,d were analyzed by two-way analysis of variance with the post hoc Bonferroni multiple comparison test. Data in c,g were analyzed by one-way analysis of variance with the post hoc Tukey’s multiple comparison test. Data in e were analyzed by two-tailed Mann Whitney’s test, data in f were analyzed by two-tailed Student’s t test. n.s., not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Fig. 6 Genetic inactivation of Clec-2 or podoplanin does not protect from CD-HFD-induced NASH.
a,b, Body weight (a) and ALT levels (b) (ND n = 5 mice, CD-HFD n = 10, CD-HFD/Clec-2 n = 9 or CD-HFD/Pdpn–/–n= 3). (c) H&E staining and (d) NAS evaluation (ND n = 5 mice, CD-HFD n = 4, CD-HFD/Clec-2–/–n= 4 or CD-HFD/Pdpn–/–n= 3), scale bar: 100 µm. All data are shown as mean ± s.e.m. and analyzed by one-way analysis of variance with the post hoc Tukey’s multiple comparison test.
Extended Data Fig. 7 MAdCAM-1 has an important role in the platelets recruitment to the liver during NASH development.
a, H&E, CD42b (ND n = 12 mice, WD-HTF n = 17 mice, WD-HTF/MAdCAM-1–/– n = 6 mice, WD-HTF/L-sel–/– n = 11 mice, WD-HTF/Beta7–/– n = 8 mice, WD-HTF/L-sel–/–/Beta7–/– n = 11 mice), CD3 (ND n = 5 mice, WD-HTF n = 8 mice, WD-HTF/MAdCAM-1–/– n = 6 mice, WD-HTF/L-sel–/– n = 5 mice, WD-HTF/Beta7–/– n = 4 mice, WD-HTF/L-sel–/–Beta7–/– n = 4 mice) and F4/80 (ND n = 4 mice, WD-HTF n = 5 mice, WD-HTF/MAdCAM-1–/– n = 5 mice, WD-HTF/-sel–/– n = 4 mice, WD-HTF/Beta7–/– n = 6 mice, WD-HTF/L-sel–/–/Beta7–/– n = 4MAdCAM-1–/– mice) stains and (b) quantification of IHC of livers of mice mentioned in a, scale bar: 50 µm. All data are shown as mean ± s.e.m. Data in b were analyzed by one-way analysis of variance with the post hoc Tukey’s multiple comparison test.
Extended Data Fig. 8 Genetic inactivation of P-selectin does not prevent NASH development.
a,b, Body weight development (ND n = 4 mice, CD-HFD n = 3 mice, CD-HFD/P-sel–/–n = 7 mice) (a) and ALT and AST levels of 6 months ND, CD-HFD or CD-HFD/P-sel–/– mice (ND n = 4 mice, CD-HFD n = 5 mice, CD-HFD/P-sel –/–n = 9 mice). Statistics: ND vs CD-HFD (black asterisks), ND vs CD-HFD/P-sel–/– (violet asterisks). c, IPGTT (ND n = 4 mice, CD-HFD n = 5 mice, CD-HFD/ n = 5 mice). Statistics: ND vs CD-HFD (black asterisks), CD-HFD vs CD-HFD/P-sel–/– (violet asterisks). d, NAS evaluation (CD-HFD n = 8 mice, CD-HFD/ P-sel–/–n = 4 mice). scale bar: 100 µm in 10×, 50 µm in 20× . e, Representative CD3, F4/80, MHCII and Ly6G stainings (CD-HFD n = 8 mice, CD-HFD/P-sel–/–n = 4 mice), scale bar: 50 µm. All data are shown as mean ± s.e.m. Data in a and c were analyzed by two-way analysis of variance with the post hoc Bonferroni multiple comparison test. Data in b were analyzed by one-way analysis of variance with the post hoc Tukey’s multiple comparison test. Data in d were analyzed by two-tailed Student’s t test. n.s., not significant *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Fig. 9 vWF–/– mice show steatosis, liver damage and conditions associated with NASH.
a, Body weight development of 6 months ND-, CD-HFD- or CD-HFD/vWF-fed mice. Statistics: (n = 6 mice/group). Statistics: ND vs CD-HFD (black asterisks), ND vs CD-HFD/vWF–/– (red asterisks). b, ALT (ND n = 4 mice, CD-HFD n = 3 mice, CD-HFD/vWF–/–n = 3 mice), liver triglycerides (ND n = 5 mice, CD-HFD n = 4 mice, CD-HFD/vWF–/–n = 5 mice) and serum cholesterol levels (ND n = 4 mice, CD-HFD n = 4 mice, CD-HFD/vWF–/–n= 3 mice). c, IPGTT (ND n = 5 mice, CD-HFD n = 5 mice, CD-HFD/vWF–/– = 3 mice). Statistics: ND vs CD-HFD (black asterisks), ND vs CD-HFD/vWF–/– (red asterisks). d, Real-time qPCR analysis for genes involved in lipid metabolism/β-oxidation (ND n = 2 mice, CD-HFD n = 4 mice, CD-HFD/ vWF–/–n= 4 mice). e, H&E with enlarged hepatocytes (asterisks) and (f) evaluation of NAS (CD-HFD n = 9 mice, CD-HFD/ vWF–/–n= 4 mice) scale bar: 100 µm in 10X and 50 µm in 20×. g, Sudan red staining and quantification of mice shown in a (n = 2–3 fields/mouse: n = 3 mice/group), scale bar: 100 µm. All data are shown as mean ± s.e.m. Data in a,c were analyzed by two-way analysis of variance with the post hoc Bonferroni multiple comparison test. Data in b,g were analyzed by one-way analysis of variance with the post hoc Tukey’s multiple comparison test. Data in d were analyzed by two-tailed Mann-Whitney test. Data in f were analyzed by two-tailed Student’s t test. n.s., not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Fig. 10 Mice lacking Mac-1 show signs of liver injury and develop NASH upon feeding CD-HFD.
a, CD42b staining and quantification of 6 months CD-HFD or CD-HFD/Mac-1–/–-fed mice (CD-HFD n = 5 mice, Mac-1/CD-HFD n = 4 mice), scale bar: 50 µm (b) Body weight (ND n = 5 mice, CD-HFD n = 8 mice, CD-HFD/Mac-1–/–n = 11 mice), (c) ALT, and AST of 6 months ND, CD-HFD or CD-HFD/Mac-1–/–-fed mice (CD-HFD n = 5 mice, CD-HFD/Mac-1–/–n = 12 mice) (d) NAS evaluation of mice shown in a (CD-HFD n = 9 mice, CD-HFD/-Mac-1–/–n = 11 mice) scale bar: 100 µm in 10× and 50 µm in 20×. e, Representative CD3+, F4/80+, MHCII+ and Ly6G+ staining (ND n = 5 mice, CD-HFD n = 8 mice, CD-HFD/Mac-1–/–n = 11 mice) and arrows indicate cell/cell aggregates, scale bar: 50 µm. All data are shown as mean ± s.e.m. Data in a,d were analyzed by two-tailed Student’s t test. Data in b,c were analyzed by one-way analysis of variance with the post hoc Tukey’s multiple comparison test.
Supplementary information
Supplementary information
Supplementary Figures 1-25, Supplementary Tables 1-4.
Source data
Source Data Fig. 3
Western blot gels.
Rights and permissions
About this article
Cite this article
Malehmir, M., Pfister, D., Gallage, S. et al. Platelet GPIbα is a mediator and potential interventional target for NASH and subsequent liver cancer. Nat Med 25, 641–655 (2019). https://doi.org/10.1038/s41591-019-0379-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-019-0379-5
This article is cited by
-
Nonalcoholic steatohepatitis-related hepatocellular carcinoma: pathogenesis and treatment
Nature Reviews Gastroenterology & Hepatology (2023)
-
Immunology of human fibrosis
Nature Immunology (2023)
-
Gut-derived low-grade endotoxaemia, atherothrombosis and cardiovascular disease
Nature Reviews Cardiology (2023)
-
Regular Aspirin Use Is Associated with a Reduced Risk of Hepatocellular Carcinoma (HCC) in Chronic Liver Disease: a Systematic Review and Meta-analysis
Journal of Gastrointestinal Cancer (2023)
-
The GPIb-IX complex on platelets: insight into its novel physiological functions affecting immune surveillance, hepatic thrombopoietin generation, platelet clearance and its relevance for cancer development and metastasis
Experimental Hematology & Oncology (2022)