Abstract
Advances in genomics and computing are transforming the capacity for the characterization of biological systems, and researchers are now poised for a precision-focused transformation in the way they prepare for, and respond to, infectious diseases. This includes the use of genome-based approaches to inform molecular diagnosis and individual-level treatment regimens. In addition, advances in the speed and granularity of pathogen genome generation have improved the capability to track and understand pathogen transmission, leading to potential improvements in the design and implementation of population-level public health interventions. In this Perspective, we outline several trends that are driving the development of precision epidemiology of infectious disease and their implications for scientists’ ability to respond to outbreaks.
Similar content being viewed by others
Main
Over the last decade, advances in genomics have driven innovations in medicine on multiple fronts. Gene sequencing, genotyping arrays, and the subsequent development of high-throughput genomics have led to detailed catalogs of human genetic variation. The completion of the Human Genome1,2, HapMap3, and the 1000 Genomes Project4 have facilitated the development of the promising field of ‘precision medicine’ and spurred the creation of large-scale initiatives, such as the UK Biobank (http://www.ukbiobank.ac.uk/) and the US-based All of Us Research Program (https://allofus.nih.gov/). These projects aim to use the power of genomics and other technologies to advance human medicine beyond interventions based on population-level averages towards personalized treatment tailored for each individual5.
Genomic technologies are also transforming another area of human health—response with precision to infectious diseases6. The world is increasingly interconnected, which, in part, is why in recent years there have been several large-scale infectious disease epidemics, often from unexpected sources, including SARS and MERS coronaviruses7,8, H1N1/A influenza virus9, Ebola virus10,11, and Zika virus12. During many of these outbreaks, sequencing of virus genomes directly from infected individuals has helped to accurately elucidate the source, timing, transmission, and spread of disease. This new field of inquiry has been termed ‘genomic epidemiology’6. During the 2013–2016 Ebola epidemic in West Africa, for example, more than 1,600 patients with Ebola (>5% of confirmed cases) had virus genomes sequenced from their blood, and the resulting data provided valuable insights into how the epidemic started, spread, and evolved11,13,14.
Epidemiological approaches to infectious disease control have traditionally relied on case (incidence) data and interview-based contact tracing to estimate key epidemic parameters (for example, basic and effective reproduction numbers, incubation period, serial interval) and to reconstruct transmission chains. These data, however, can be limited by incomplete case reporting due to the labor-intensive nature of contact tracing or uncertain reporting due to the use of clinical symptoms to identify cases.
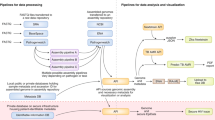
Although these traditional data sources still play critical roles in informing outbreak interventions, high-throughput and near-real-time pathogen genome sequencing is transforming infectious disease epidemiology6,11,12. By increasing both the scale and resolution of inference, genomic technologies are enabling a more targeted approach to infectious disease control at both the individual and population level, which we refer to, collectively, as ‘precision epidemiology’ (Table 1). We will briefly outline how genomic technologies are enabling precision epidemiology by allowing the design of better intervention strategies for individual patients and for affected populations as a whole (Fig. 1).
Technological advances are enabling the broad application of pathogen genome sequencing for our response to outbreaks of infectious disease. Whole-genome sequencing of many pathogens can now be done directly from clinical samples and in near real time during an outbreak. By analyzing these genomes and their metadata in the context of other sequences generated from the same outbreak, as well as previously characterized variants, researchers can inform individual- and population-level intervention strategies to minimize the burden of infectious diseases. We term the collective approach—sequencing, analysis, and response—as precision epidemiology.
Precision epidemiology in the clinic
The driving principle behind precision medicine is that one size does not, in fact, fit all15. To date, the field has primarily focused on the use of patients’ own genomic information to make personalized decisions about disease treatment5. During infectious disease outbreaks, however, genomic sequence information from the pathogen is arguably more important than an individual’s genomic data for designing appropriate treatment and intervention strategies16.
The practice of utilizing pathogen genotypic information for the diagnosis and treatment of infectious diseases is not new, but technological advances, most notably in the targeted enrichment of pathogen nucleic acids17,18,19 and next-generation sequencing20, have greatly improved the prospect of broadly applying this approach in the clinic. In the past, practical applications of pathogen genotyping were limited by the slow pace of sequencing and its focus only on specific genes—or even portions of genes. Today, in contrast, researchers can characterize entire viral and bacterial genomes from infected individuals in near real time6. Given enough sequence coverage, they can also characterize minor genetic variants in pathogen genomes present within an individual patient, which can be critically relevant in directing clinical care21,22.
Although not typically presented as precision medicine, pathogen genomic information has been used successfully to assess drug sensitivity and/or resistance on a patient-by-patient basis for several significant human pathogens, including HIV23, influenza virus21, and Mycobacterium tuberculosis24. This information can be used—in a manner analogous to human genotypes—to guide the design of individualized drug regimens (for example, antibiotics and antivirals) (Fig. 1). Applying genomic technologies during the development and usage of immunotherapeutics (for example, monoclonal antibody cocktails25) and vaccines can also provide insights into pathogen strategies for immune response evasion26,27 and mechanisms of virulence28,29. By characterizing longitudinal samples from the same patients, pathogen sequencing also provides the potential for identifying genetic components involved in driving disease progression, thus providing novel drug targets30.
Point-of-care molecular tests tailored to individual pathogens have dramatically increased the speed and specificity of infectious disease diagnosis, though there is still considerable room for improvements in sensitivity31. One advantage of genomic approaches is that molecular diagnostics can be modified in light of pathogen sequence information generated during an outbreak6. This, for example, was achieved during the 2013–2016 Ebola epidemic, when rapidly generated virus genome sequences were used to update PCR-based diagnostics so that they more closely matched the Makona variant of Ebola virus responsible for the epidemic32.
In addition to the utility of genomic technologies for improving traditional diagnostic tests, metagenomic next-generation sequencing—in which all genomic information, including microbial material, is sequenced in an untargeted manner—holds great promise as a general approach for the detection and characterization of pathogens without the need for a priori knowledge of the potential causative agent33,34. Because metagenomic approaches do not target particular pathogens, they are equally applicable to the detection of expected pathogens as they are to the detection of novel pathogens—such as the emergences of SARS7 and MERS8—or to the detection of known pathogens in new places, as was illustrated by Ebola virus in West Africa during the 2013–2016 epidemic14. The combination of highly multiplexed target capture and next-generation sequencing is particularly promising, as it increases both sensitivity and specificity. Such an approach is feasible because it is now possible to multiplex millions of individual pathogen-specific probes, each of which can enrich for highly divergent nucleic acids (up to ~40% divergence)19.
Precision epidemiology informs outbreak response
Pathogen genomes can also be used to inform population-level intervention strategies for infectious disease outbreaks. In contrast to the design of individual-level treatment strategies, in which the functional roles of host and/or pathogen mutations are critical, outbreak-scale genomic analyses use pathogen mutations as markers of transmission events. Genomic epidemiology exploits the rapid evolution of pathogens, which often accumulate mutations on the same timescale as their epidemiological spread35, to reconstruct outbreak dynamics from genomic data. With sufficient sampling, relevant metadata (such as location and date) and an appropriate statistical framework, pathogen genomes can reveal patterns of epidemic transmission at a fine-scale resolution, thus enabling the design of targeted interventions that are more precise than those based on traditional epidemiological data alone.
One application of precision epidemiology during outbreaks is the identification of causal pathogens and their modes of transmission. Large-scale virus genome sequencing efforts during the 2013–2016 Ebola epidemic, for example, showed that it resulted from a single cross-species ‘spillover’ event of Zaire ebolavirus, from an animal reservoir to humans, followed by sustained human-to-human transmission11. However, while human-to-human transmission typically occurs through direct contact with bodily fluids from a symptomatic individual, genomic epidemiology also demonstrated the potential for sexual transmission of Ebola virus from persistently infected asymptomatic individuals36. This mode of dissemination played a critical role in prolonging the Ebola epidemic in West Africa, and as a result of genomic studies, the World Health Organization (WHO) made an immediate change to their guidance for Ebola survivors and reccomended repeated diagnostic characterization of semen samples prior to two consecutive negative results37. In contrast, genomic epidemiological studies of Lassa fever, which is endemic in West Africa38, showed that human cases of Lassa fever are the result of multiple independent spillovers from a Mastomys natalensis rodent reservoir, with limited human-to-human transmission38,39.
One of the most advanced population-level applications of precision epidemiology is food safety, where it is used for pathogen identification and source attribution. Genome sequencing of foodborne bacterial pathogens now forms part of many surveillance systems, and outbreak investigations in the United States are routinely performed by the Food and Drug Administration’s GenomeTrackr Network. In recent years, this network has grown into an international collaboration among 63 government, private, and academic research laboratories40,41. Through near-real-time genome sequencing and public data deposition of clinical, environmental, and food-related bacterial isolates, this network is streamlining the process of recognizing, investigating, and reducing the impact of foodborne disease outbreaks42,43. The success of this approach was demonstrated recently through a broad investigation of several foodborne Listeria monocytogenes outbreaks across the United States44.
Phylogenetic analysis of pathogen genomes can also be used to elucidate the spatial and temporal scales of transmission, which are critical for the design of effective public health interventions. HIV sequences, for example, have been used to reconstruct transmission networks in detail, with the goal of focusing the use of antiretroviral drugs, along with screening and prevention education messages, in a targeted manner to interrupt community spread45,46. Likewise, Zika virus genomes have been used to determine the relative contributions to epidemic growth of local vector-borne transmission versus repeated reintroductions from travelers in sustaining Zika outbreaks in the Americas47,48. Phylogenetic investigations have also been critical for disentangling the roles of community- and hospital-based transmission of bacterial pathogens49. In one example, whole-genome sequences of methicillin-resistant Staphylococcus aureus (MRSA) indicated that a persistently infected healthcare worker in Cambridge, UK likely played a key role in sustaining transmission within a particular hospital unit50. This analysis directly led to infection control interventions, including targeted pathogen decolonization efforts.
Genomically informed transmission trees are also used to directly estimate key epidemic parameters (such as the basic and effective reproduction numbers of an outbreak), either independently or in combination with incidence data51. Such analyses can provide rapid estimates of pandemic potential and are used to evaluate the effectiveness of interventions51,52. Genomic data can even provide information on within-outbreak population structure (that is, differences in transmission dynamics between geographic locations or risk groups)53 and the proportion of unreported cases54.
Finally, sequencing allows us to monitor genetic changes over time in pathogen populations, an understanding of which is critical for the design of effective diagnostics and countermeasures. Vaccines, for example, are our primary line of defense against seasonal influenza. However, influenza viruses evolve quickly to evade immune responses to previously circulating variants or prior vaccinations. Genetic sequencing and large-scale bioinformatic analysis provide powerful tools for tracking the evolution of influenza viruses in real time55 and for predicting the strains likely to be most prevalent each year. The seasonal influenza vaccine can then be regularly updated to reflect projected changes in the global population of influenza strains56.
Challenges for precision epidemiology during outbreaks
Advances in sequencing technologies are enabling the development and use of innovative genomic approaches for the treatment and prevention of infectious diseases. Adoption of genomic epidemiology into effective outbreak responses, however, will require the establishment of improved mechanisms for coordination between academic researchers and public health agencies. This includes changes to research practice regarding the benefits for rapid and open sharing of data and results as well as a focus on building capacity for sequencing and analysis within public health agencies and the regions most severely impacted by infectious disease57,58.
Comprehensive and carefully organized sampling of pathogen genomes from patients along with rich sets of metadata (Box 1) are required to improve the accuracy and resolution of outbreak transmission patterns reconstructed using genomic epidemiology. Sampling is typically performed or coordinated by local hospitals and departments of health, national entities such as the US Centers for Disease Control and Prevention (CDC), or international groups like the World Health Organization (WHO) and Médecins Sans Frontières. Expertise in genome sequencing, bioinformatics, and phylogenetic analysis, in contrast, is typically concentrated within academic and government research laboratories. Therefore, at this point in time, for precision epidemiology to be successfully implemented, it is critical that researchers and public health agencies work together in close coordination. Such collaborations were critical during responses to the recent Ebola and Zika epidemics; however, the approach to establishing these partnerships was largely unsystematic and, in many cases, delayed because of the need to establish relationships during the course of public health emergencies59.
One important approach to accelerating responses in the future is to build genome sequencing and analysis capabilities within public health agencies and hospitals as well as in developing countries disproportionately impacted by infectious disease outbreaks. Several such efforts are currently underway, including the Association of Publich Health Laboratories (APHL)–CDC bioinformatics fellowship program (https://www.aphl.org/fellowships/pages/bioinformatics.aspx) and the H3Africa initiative, which is backed by the US National Institutes of Health and the UK Wellcome Trust60. Genomics programs within public health agencies and at individual hospitals would streamline the process of integrating genomic data into outbreak response efforts. Genomic epidemiology, however, is a rapidly evolving field with a strong theoretical foundation, and owing to differences in priorities, academic research groups will likely continue to be at the forefront of tool development and implementation. Therefore, it is imperative that researchers develop a framework of norms and rules governing research conduct during and between outbreaks61, establish diverse networks of technical response teams, and produce action plans. This framework needs to be implemented in advance of an outbreak and coordinated through international organizations, like the WHO, and oversight committees within the United Nations59.
It is critical that data and analyses are shared openly during infectious disease outbreaks to ensure the most comprehensive and efficient response possible while ethical constraints also receive close attention. This includes the public release of raw genome sequence data as well as analysis results, which should be provided in a format that conveys to nonspecialists the complexities and uncertainties associated with interpretation. Further development of portable instruments6 for in-country sequencing and online analysis platforms62,63 will continue to advance the rapid generation and open dissemination of data, analyses, and actionable insights. However, concerns regarding the perceived career benefits of slower or more limited public access to outbreak data remain a barrier to open science within the research community. Despite this, there are signs of progress. During recent outbreaks, many researchers made data and analyses available and participated in open discussions via online depositories and forums, such as GitHub and Virological.org, with complete manuscripts often made available prior to publication via preprint servers such as the bioRxiv64. We hope that the successes of the research collaborations that followed this approach will help to increase participation in the future. These movements towards making outbreak data more openly available are also supported by several major public health agencies, including the WHO, which recently called for data relevant to public health emergencies to be distributed immediately and freely upon generation65,66.
With the current capabilities, cost, and speed of sequencing technologies, the field has finally reached a point where rapid genomic surveillance and analysis can start to become a standard part of the response to infectious disease outbreaks. Just as broadscale human genome sequencing revolutionized the treatment of many noncommunicable diseases, pathogen genome data are poised to drive a similar revolution in the response to infectious diseases.
References
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Venter, J. C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001).
International HapMap Consortium. The International HapMap Project. Nature 426, 789–796 (2003).
1000 Genomes Project Consortium et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Torkamani, A., Andersen, K. G., Steinhubl, S. R. & Topol, E. J. High-definition medicine. Cell 170, 828–843 (2017).
Gardy, J. L. & Loman, N. J. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat. Rev. Genet. 19, 9–20 (2018).
Ksiazek, T. G. et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1953–1966 (2003).
Zaki, A. M., van Boheemen, S., Bestebroer, T. M., Osterhaus, A. D. M. E. & Fouchier, R. A. M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820 (2012).
Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 360, 2605–2615 (2009).
Baize, S. et al. Emergence of Zaire Ebola virus disease in Guinea. N. Engl. J. Med. 371, 1418–1425 (2014).
Holmes, E. C., Dudas, G., Rambaut, A. & Andersen, K. G. The evolution of Ebola virus: insights from the 2013-2016 epidemic. Nature 538, 193–200 2016).
Grubaugh, N. D., Faria, N. R., Andersen, K. G. & Pybus, O. G. Genomic insights into Zika virus emergence and spread. Cell 172, 1160–1162 2018).
Dudas, G. et al. Virus genomes reveal factors that spread and sustained the Ebola epidemic. Nature 544, 309–315 (2017).
Gire, S. K. et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 345, 1369–1372 (2014).
Collins, F. S. & Varmus, H. A new initiative on precision medicine. N. Engl. J. Med. 372, 793–795 (2015).
Bissonnette, L. & Bergeron, M. G. Infectious disease management through point-of-care personalized medicine molecular diagnostic technologies. J. Pers. Med. 2, 50–70 (2012).
Matranga, C. B.et al. Enhanced methods for unbiased deep sequencing of Lassa and Ebola RNA viruses from clinical and biological samples. Genome Biol. 15, 519 (2014).
Quick, J. et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc. 12, 1261–1276 (2017).
Wylie, T. N., Wylie, K. M., Herter, B. N. & Storch, G. A. Enhanced virome sequencing using targeted sequence capture. Genome Res. 25, 1910–1920 (2015).
Shendure, J. & Ji, H. Next-generation DNA sequencing. Nat. Biotechnol. 26, 1135–1145 (2008).
Kiso, M. et al. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet 364, 759–765 (2004).
Simen, B. B. et al. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J. Infect. Dis. 199, 693–701 (2009).
Perrin, L. HIV treatment failure: testing for HIV resistance in clinical practice. Science 280, 1871–1873 (1998).
Schön, T.et al. Mycobacterium tuberculosis drug-resistance testing: challenges, recent developments and perspectives. Clin. Microbiol. Infect. 23, 154–160 2017).
Both, L. et al. Monoclonal antibodies for prophylactic and therapeutic use against viral infections. Vaccine 31, 1553–1559 (2013).
Kugelman, J. R. et al. Emergence of ebola virus escape variants in infected nonhuman primates treated with the MB-003 antibody cocktail. Cell Rep. 12, 2111–2120 (2015).
Doud, M. B., Hensley, S. E. & Bloom, J. D. Complete mapping of viral escape from neutralizing antibodies. PLoS. Pathog. 13, e1006271 (2017).
Cann, A. J. et al. Reversion to neurovirulence of the live-attenuated Sabin type 3 oral poliovirus vaccine. Nucleic Acids Res. 12, 7787–7792 (1984).
Stern, A. et al. The Evolutionary Pathway to Virulence of an RNA Virus. Cell 169, 35–46.e19 (2017).
Kugelman, J. R. et al. Evaluation of the potential impact of Ebola virus genomic drift on the efficacy of sequence-based candidate therapeutics. MBio 6, e02227-14 (2015).
Kozel, T. R. & Burnham-Marusich, A. R. Point-of-care testing for infectious diseases: past, present, and future. J. Clin. Microbiol. 55, 2313–2320 (2017).
Sozhamannan, S. et al. Evaluation of signature erosion in ebola virus due to genomic drift and its impact on the performance of diagnostic assays. Viruses 7, 3130–3154 (2015).
Greninger, A. L. et al. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med. 7, 99 (2015).
Miller, S. et al. Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal Ffuid. Preprint at https://doi.org/10.1101/330381 (2018).
Biek, R., Pybus, O. G., Lloyd-Smith, J. O. & Didelot, X. Measurably evolving pathogens in the genomic era. Trends. Ecol. Evol. 30, 306–313 (2015).
Mate, S. E. et al. Molecular evidence of sexual transmission of ebola virus. N. Engl. J. Med. 373, 2448–2454 (2015).
Butler, D. What first case of sexually transmitted Ebola means for public health. Nature News https://doi.org/10.1038/nature.2015.18584 (2015).
Andersen, K. G. et al. Clinical sequencing uncovers origins and evolution of lassa virus. Cell 162, 738–750 (2015).
Siddle, K. J. et al. Genomic analysis of Lassa virus during an increase in cases in Nigeria in 2018. N. Engl. J. Med. 379,1745–1753 (2018).
Timme, R. E. et al. GenomeTrakr proficiency testing for foodborne pathogen surveillance: an exercise from 2015. Microb Genom 4, https://doi.org/10.1099/mgen.0.000185 (2018).
Center for Food Safety & Nutrition, A. Whole Genome Sequencing Program—GenomeTrakr Network https://www.fda.gov/food/foodscienceresearch/wholegenomesequencingprogramwgs/ucm363134.htm (2018).
Allard, M. W. et al. Genomics of foodborne pathogens for microbial food safety. Curr. Opin. Biotechnol. 49, 224–229 (2018).
Stevens, E. L. et al. The public health impact of a publically available, environmental database of microbial genomes. Front. Microbiol. 8, 808 (2017).
Jackson, B. R. et al. Implementation of nationwide real-time whole-genome sequencing to enhance listeriosis outbreak detection and investigation. Clin. Infect. Dis. 63, 380–386 (2016).
Grabowski, M. K. et al. The role of viral introductions in sustaining community-based HIV epidemics in rural Uganda: evidence from spatial clustering, phylogenetics, and egocentric transmission models. PLoS Med. 11, e1001610 (2014).
Little, S. J. et al. Using HIV networks to inform real time prevention interventions. PLoS ONE. 9, e98443 (2014).
Grubaugh, N. D. et al. Genomic epidemiology reveals multiple introductions of Zika virus into the United States. Nature 546, 401–405 (2017).
Faria, N. R. et al. Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature 506, 406–410 (2017).
Popovich, K. J. & Snitkin, E. S. Whole genome sequencing-implications for infection prevention and outbreak investigations. Curr. Infect. Dis. Rep. 19, 15 (2017).
Harris, S. R. et al. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet. Infect. Dis. 13, 130–136 (2013).
du Plessis, L. & Stadler, T. Getting to the root of epidemic spread with phylodynamic analysis of genomic data. Trends Microbiol. 23, 383–386 (2015).
Fraser, C. et al. Pandemic potential of a strain of influenza A (H1N1): early findings. Science 324, 1557–1561 (2009).
Kühnert, D., Stadler, T., Vaughan, T. G. & Drummond, A. J. Phylodynamics with migration: a computational framework to quantify population structure from genomic data. Mol. Biol. Evol. 33, 2102–2116 (2016).
Stadler, T., Kühnert, D., Rasmussen, D. A. & du Plessis, L. Insights into the early epidemic spread of Ebola in Sierra Leone provided by viral sequence data. PLoS Curr. 6, http://doi.org/10.1371/currents.outbreaks.02bc6d927ecee7bbd33532ec8ba6a25f (2014).
Neher, R. A. & Bedford, T. nextflu: real-time tracking of seasonal influenza virus evolution in humans. Bioinformatics 31, 3546–3548 (2015).
Ampofo, W. K. et al. Strengthening the influenza vaccine virus selection and development process: Report of the 3rd WHO Informal Consultation for Improving Influenza Vaccine Virus Selection held at WHO headquarters, Geneva, Switzerland, 1-3 April 2014. Vaccine 33, 4368–4382 (2015).
Yozwiak, N. L. et al. Roots, not parachutes: research collaborations combat outbreaks. Cell 166, 5–8 (2016).
Yozwiak, N. L., Schaffner, S. F. & Sabeti, P. C. Data sharing: make outbreak research open access. Nature 518, 477–479 (2015).
Moon, S. et al. Will Ebola change the game? Ten essential reforms before the next pandemic. The report of the Harvard-LSHTM independent panel on the global response to ebola. Lancet 386, 2204–2221 (2015).
H3Africa Consortium. et al. Research capacity. Enabling the genomic revolution in Africa. Science 344, 1346–1348 (2014).
World Health Organization. Guidance for Managing Ethical Issues in Infectious Disease Outbreaks (World Health Organization, 2016).
Hadfield, J. et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34, 4121–4123 (2018).
Brownstein, J. S. & Freifeld, C. C. HealthMap: the development of automated real-time internet surveillance for epidemic intelligence. Euro. Surveill. 12, E071129.5 (2007).
Johansson, M. A., Reich, N. G., Meyers, L. A. & Lipsitch, M. Preprints: an underutilized mechanism to accelerate outbreak science. PLoS. Med. 15, e1002549 (2018).
WHO. Policy statement on data sharing by WHO in the context of public health emergencies (as of 13 April 2016). Wkly. Epidemiol. Rec. 91, 237–240 (2016).
WHO. R&D Blueprint Meeting on Pathogen Genetic Sequence Data (GSD) Sharing in the Context of Public Health Emergencies, 28-29 September 2017 (WHO, 2017).
Christie, A. et al. Possible sexual transmission of Ebola virus - Liberia, 2015. MMWR. Morb. Mortal. Wkly. Rep. 64, 479–481 (2015).
Poon, A. F. Y. et al. Near real-time monitoring of HIV transmission hotspots from routine HIV genotyping: an implementation case study. Lancet HIV 3, e231–e238 (2016).
Eyre, D. W. et al. A Candida Auris outbreak and its control in an intensive care setting. N. Engl. J. Med. 379, 1322–1331 (2018).
Faria, N. R. et al. Genomic and epidemiological monitoring of yellow fever virus transmission potential. Science 361, 894–899 (2018).
Briese, T. et al. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS. Pathog. 5, e1000455 (2009).
Neher, R. A., Bedford, T., Daniels, R. S., Russell, C. A. & Shraiman, B. I. Prediction, dynamics, and visualization of antigenic phenotypes of seasonal influenza viruses. Proc. Natl Acad. Sci. USA 113, E1701–E1709 (2016).
Grad, Y. H. et al. Genomic epidemiology of the Escherichia coli O104:H4 outbreaks in Europe, 2011. Proc. Natl Acad. Sci. USA 109, 3065–3070 (2012).
Robinson, P. N. Deep phenotyping for precision medicine. Hum. Mutat. 33, 777–780 (2012).
Pan, H. et al. Using PhenX measures to identify opportunities for cross-study analysis. Hum. Mutat. 33, 849–857 (2012).
Robinson, P. N. et al. The human phenotype ontology: a tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 83, 610–615 (2008).
Bogue, M. A. et al. Mouse phenome database: an integrative database and analysis suite for curated empirical phenotype data from laboratory mice. Nucleic Acids Res. 46, D843–D850 (2018).
Cheng, K. C., Xin, X., Clark, D. P. & La Riviere, P. Whole-animal imaging, gene function, and the Zebrafish Phenome Project. Curr. Opin. Genet. Dev. 21, 620–629 (2011).
Alexandrov, V. et al. Large-scale phenome analysis defines a behavioral signature for Huntington’s disease genotype in mice. Nat. Biotechnol. 34, 838–844 (2016).
Zola, S. M., Manzanares, C. M., Clopton, P., Lah, J. J. & Levey, A. I. A behavioral task predicts conversion to mild cognitive impairment and Alzheimer’s disease. Am. J. Alzheimers Dis. Other Demen. 28, 179–184 (2013).
Harrington, J., Schramm, P. J., Davies, C. R. & Lee-Chiong, T. L. Jr. An electrocardiogram-based analysis evaluating sleep quality in patients with obstructive sleep apnea. Sleep. Breath. 17, 1071–1078 (2013).
Boulding, H. & Webber, C. Large-scale objective association of mouse phenotypes with human symptoms through structural variation identified in patients with developmental disorders. Hum. Mutat. 33, 874–883 (2012).
Brownstein, J. S., Freifeld, C. C. & Madoff, L. C. Digital disease detection — harnessing the web for public health surveillance. N. Engl. J. Med. 360, 2153–2157 (2009).
Bourgeois, F. T. et al. The value of patient self-report for disease surveillance. J. Am. Med. Inform. Assoc. 14, 765–771 (2007).
Milinovich, G. J. et al. Using internet search queries for infectious disease surveillance: screening diseases for suitability. BMC. Infect. Dis. 14, 690 (2014).
Charles-Smith, L. E. et al. Using social media for actionable disease surveillance and outbreak management: a systematic literature review. PLoS. ONE. 10, e0139701 (2015).
Nsoesie, E. O. et al. Social media as a sentinel for disease surveillance: what does sociodemographic status have to do with It? PLoS Curr. 8, http://doi.org/ecurrents.outbreaks.cc09a42586e16dc7dd62813b7ee5d6b6 (2016).
Acknowledgements
O.G.P. is supported by the Oxford Martin School. K.G.A. is a Pew Biomedical Scholar and is supported by NIH NCATS CTSA UL1TR001114, NIAID HHSN272201400048C, NIAID R21AI137690, NIAID U19AI135995, and The Ray Thomas Foundation. J.T.L. is supported by the State of Arizona Technology and Research Initiative Fund (TRIF), administered by the Arizona Board of Regents, through Northern Arizona University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ladner, J.T., Grubaugh, N.D., Pybus, O.G. et al. Precision epidemiology for infectious disease control. Nat Med 25, 206–211 (2019). https://doi.org/10.1038/s41591-019-0345-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-019-0345-2
This article is cited by
-
Inferring bacterial transmission dynamics using deep sequencing genomic surveillance data
Nature Communications (2023)
-
The importance of equally accessible genomic surveillance in the age of pandemics
Biologia Futura (2023)
-
VirStrain: a strain identification tool for RNA viruses
Genome Biology (2022)
-
Genomic surveillance of Rift Valley fever virus: from sequencing to lineage assignment
BMC Genomics (2022)
-
AusTrakka: Fast-tracking nationalized genomics surveillance in response to the COVID-19 pandemic
Nature Communications (2022)