Abstract
Apolipoprotein-E (ApoE) has been implicated in Alzheimer’s disease, atherosclerosis, and other unresolvable inflammatory conditions but a common mechanism of action remains elusive. We found in ApoE-deficient mice that oxidized lipids activated the classical complement cascade (CCC), resulting in leukocyte infiltration of the choroid plexus (ChP). All human ApoE isoforms attenuated CCC activity via high-affinity binding to the activated CCC-initiating C1q protein (KD~140–580 pM) in vitro, and C1q–ApoE complexes emerged as markers for ongoing complement activity of diseased ChPs, Aβ plaques, and atherosclerosis in vivo. C1q–ApoE complexes in human ChPs, Aβ plaques, and arteries correlated with cognitive decline and atherosclerosis, respectively. Treatment with small interfering RNA (siRNA) against C5, which is formed by all complement pathways, attenuated murine ChP inflammation, Aβ-associated microglia accumulation, and atherosclerosis. Thus, ApoE is a direct checkpoint inhibitor of unresolvable inflammation, and reducing C5 attenuates disease burden.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All microarray data can be found under GEO database accession number GSE85774 and GSE85775 for ChPs. Microarray data for aorta had been published previously with the accession number GSE40156 (ref. 57). The remaining source data for figures in the manuscript will be made available upon reasonable request.
Change history
04 February 2019
In the version of this article originally published, a sentence was erroneously included in the author contributions, and information regarding second shared authorship was missing from the author contributions. The following should not have been included in the author contributions: “C.W. and A.J.R.H. supervised the work presented in Figs. 1, 2, 5, 6; P.Z. and C.S. supervised the work presented in Figs. 3, 4.” Additionally, this sentence should have appeared at the beginning of the author contributions: “These authors contributed equally: C.W., P.F.Z., C.S., and A.J.R.H.” The errors have been corrected in the print, PDF and HTML versions of the article.
References
Bell, R. D. et al. Apolipoprotein E controls cerebrovascular integrity via cyclophilin A. Nature 485, 512–516 (2012).
Holtzman, D. M., Herz, J. & Bu, G. Apolipoprotein E and apolipoprotein E receptors: normal biology and roles in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2, a006312 (2012).
Kanekiyo, T., Xu, H. & Bu, G. ApoE and Aβ in Alzheimer’s disease: accidental encounters or partners? Neuron 81, 740–754 (2014).
Linton, M. F., Atkinson, J. B. & Fazio, S. Prevention of atherosclerosis in apolipoprotein E-deficient mice by bone marrow transplantation. Science 267, 1034–1037 (1995).
Mahley, R. W., Weisgraber, K. H. & Huang, Y. Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer’s disease to AIDS. J. Lipid Res. 50 Suppl, S183–S188 (2009).
Zlokovic, B. V. Cerebrovascular effects of apolipoprotein E: implications for Alzheimer disease. JAMA Neurol 70, 440–444 (2013).
Mahley, R. W. & Huang, Y. Apolipoprotein E sets the stage: response to injury triggers neuropathology. Neuron 76, 871–885 (2012).
Hansson, G. K. & Hermansson, A. The immune system in atherosclerosis. Nat. Immunol. 12, 204–212 (2011).
Que, X. et al. Oxidized phospholipids are proinflammatory and proatherogenic in hypercholesterolaemic mice. Nature 558, 301–306 (2018).
Goldmann, T. et al. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat. Immunol. 17, 797–805 (2016).
Lun, M. P., Monuki, E. S. & Lehtinen, M. K. Development and functions of the choroid plexus-cerebrospinal fluid system. Nat. Rev. Neurosci. 16, 445–457 (2015).
Moore, G. R. et al. Complement and humoral adaptive immunity in the human choroid plexus: roles for stromal concretions, basement membranes, and epithelium. J. Neuropathol. Exp. Neurol. 75, 415–428 (2016).
Ransohoff, R. M. & Engelhardt, B. The anatomical and cellular basis of immune surveillance in the central nervous system. Nat. Rev. Immunol. 12, 623–635 (2012).
Schwartz, M. & Baruch, K. The resolution of neuroinflammation in neurodegeneration: leukocyte recruitment via the choroid plexus. EMBO J. 33, 7–22 (2014).
Baruch, K. et al. Aging. Aging-induced type I interferon response at the choroid plexus negatively affects brain function. Science 346, 89–93 (2014).
Binder, C. J., Papac-Milicevic, N. & Witztum, J. L. Innate sensing of oxidation-specific epitopes in health and disease. Nat. Rev. Immunol. 16, 485–497 (2016).
Weismann, D. et al. Complement factor H binds malondialdehyde epitopes and protects from oxidative stress. Nature 478, 76–81 (2011).
Zipfel, P. F. & Skerka, C. Complement regulators and inhibitory proteins. Nat. Rev. Immunol. 9, 729–740 (2009).
Arlaud, G. J., Biro, A. & Ling, W. L. Enzymatically modified low-density lipoprotein is recognized by C1q and activates the classical complement pathway. J. Lipids 2011, 376092 (2011).
Haapasalo, K. et al. Complement factor H binds to human serum apolipoprotein E and mediates complement regulation on high density lipoprotein particles. J. Biol. Chem. 290, 28977–28987 (2015).
Wilson, C. et al. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science 252, 1817–1822 (1991).
Venkatraman Girija, U. et al. Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation. Proc. Natl Acad. Sci. USA 110, 13916–13920 (2013).
Braak, H. et al. Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol. 112, 389–404 (2006).
Thal, D. R., Rüb, U., Orantes, M. & Braak, H. Phases of Aβ-deposition in the human brain and its relevance for the development of AD. Neurology 58, 1791–1800 (2002).
Soderberg, O. et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 (2006).
Strittmatter, W. J. et al. Isoform-specific interactions of apolipoprotein E with microtubule-associated protein tau: implications for Alzheimer disease. Proc. Natl Acad. Sci. USA 91, 11183–11186 (1994).
Radde, R. et al. Aβ42-driven cerebral amyloidosis in transgenic mice reveals early and robust pathology. EMBO Rep. 7, 940–946 (2006).
Grabner, R. et al. Lymphotoxin beta receptor signaling promotes tertiary lymphoid organogenesis in the aorta adventitia of aged ApoE−/− mice. J. Exp. Med. 206, 233–248 (2009).
Stary, H. C. Natural history and histological classification of atherosclerotic lesions: an update. Arterioscler. Thromb. Vasc. Biol. 20, 1177–1178 (2000).
Hong, S. et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352, 712–716 (2016).
Stevens, B. et al. The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178 (2007).
Kolev, M., Friec, G. L. & Kemper, C. Complement - tapping into new sites and effector systems. Nat. Rev. Immunol. 14, 811–820 (2014).
Vasek, M. J. et al. A complement–microglial axis drives synapse loss during virus-induced memory impairment. Nature 534, 538–543 (2016).
Hajishengallis, G. et al. Novel mechanisms and functions of complement. Nat. Immunol. 18, 1288–1298 (2017).
Dinarello, C. A. Anti-inflammatory agents: present and future. Cell 140, 935–950 (2010).
Ricklin, D. & Lambris, J. D. Complement in immune and inflammatory disorders: pathophysiological mechanisms. J. Immunol. 190, 3831–3838 (2013).
Tabas, I. & Glass, C. K. Anti-inflammatory therapy in chronic disease: challenges and opportunities. Science 339, 166–172 (2013).
Fonseca, M. I., Zhou, J., Botto, M. & Tenner, A. J. Absence of C1q leads to less neuropathology in transgenic mouse models of Alzheimer’s disease. J. Neurosci. 24, 6457–6465 (2004).
Huang, Y. A., Zhou, B., Wernig, M. & Sudhof, T. C. ApoE2, ApoE3, and ApoE4 differentially stimulate APP transcription and Abeta secretion. Cell 168, 427–441.e421 (2017).
Shi, Y. et al. ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature 549, 523–527 (2017).
Bales, K. R. et al. Lack of apolipoprotein E dramatically reduces amyloid beta-peptide deposition. Nat. Genet. 17, 263–264 (1997).
Huynh, T. V. et al. Age-dependent effects of apoE reduction using antisense oligonucleotides in a model of beta-amyloidosis. Neuron 96, 1013–1023.e1014 (2017).
Liu, C. C. et al. ApoE4 accelerates early seeding of amyloid pathology. Neuron 96, 1024–1032.e1023 (2017).
Ulrich, J. D. et al. ApoE facilitates the microglial response to amyloid plaque pathology. J. Exp. Med. 215, 1047–1058 (2018).
Wang, C. et al. Gain of toxic apolipoprotein E4 effects in human iPSC-derived neurons is ameliorated by a small-molecule structure corrector. Nat. Med. 24, 647–657 (2018).
Tenner, A. J., Stevens, B. & Woodruff, T. M. New tricks for an ancient system: Physiological and pathological roles of complement in the CNS. Mol. Immunol. 102, 3–13 (2018).
Qiu, C. & Fratiglioni, L. A major role for cardiovascular burden in age-related cognitive decline. Nat. Rev. Cardiol. 12, 267–277 (2015).
Hofman, A. et al. Atherosclerosis, apolipoprotein E, and prevalence of dementia and Alzheimer’s disease in the Rotterdam Study. Lancet 349, 151–154 (1997).
Macedo, A. C. L. & Isaac, L. Systemic lupus erythematosus and deficiencies of early components of the complement classical pathway. Front. Immunol. 7, 55 (2016).
Zhang, B. et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 153, 707–720 (2013).
Knouff, C. et al. Apo E structure determines VLDL clearance and atherosclerosis risk in mice. J. Clin. Invest. 103, 1579–1586 (1999).
Alafuzoff, I. et al. Staging of neurofibrillary pathology in Alzheimer’s disease: a study of the BrainNet Europe Consortium. Brain. Pathol. 18, 484–496 (2008).
Alafuzoff, I. et al. Assessment of beta-amyloid deposits in human brain: a study of the BrainNet Europe Consortium. Acta Neuropathol. 117, 309–320 (2009).
Hyman, B. T. et al. National institute on aging-Alzheimer’s association guidelines for the neuropathologic assessment of Alzheimer’s disease. Alzheimers Dement. 8, 1–13 (2012).
Timaran, C. H., McKinsey, J. F., Schneider, P. A. & Littooy, F. Reporting standards for carotid interventions from the Society for Vascular Surgery. J. Vasc. Surg. 53, 1679–1695 (2011).
Abbott, A. L. et al. Systematic review of guidelines for the management of asymptomatic and symptomatic carotid stenosis. Stroke 46, 3288–3301 (2015).
Hu, D. et al. Artery Tertiary lymphoid organs control aorta immunity and protect against atherosclerosis via vascular smooth muscle cell lymphotoxin β receptors. Immunity 42, 1100–1115 (2015).
Greissel, A. et al. Alternation of histone and DNA methylation in human atherosclerotic carotid plaques. Thromb. Haemost. 114, 390–402 (2015).
Wendorff, C. et al. Carotid plaque morphology is significantly associated with sex, age, and history of neurological symptoms. Stroke 46, 3213–3219 (2015).
Franklin, K. & George, P. The Mouse Brain in Stereotaxic Coordinates, Compact 3rd edn. (Academic Press, Cambridge, MA, 2007).
Zhao, L. et al. The 5-lipoxygenase pathway promotes pathogenesis of hyperlipidemia-dependent aortic aneurysm. Nat. Med. 10, 966–973 (2004).
Haege, S. et al. CXC Chemokine receptor 7 (CXCR7) regulates CXCR4 protein expression and capillary tuft development in mouse kidney. PLoS One 7, e42814 (2012).
Yin, C. et al. Generation of aorta transcript atlases of wild-type and apolipoprotein e-null mice by laser capture microdissection-based mRNA expression microarrays. Methods Mol. Biol. 1339, 297–308 (2015).
Beer, M. et al. Laser-capture microdissection of hyperlipidemic/ApoE−/− mouse aorta atherosclerosis. Methods Mol. Biol. 755, 417–428 (2011).
Koch, T. K. et al. Staphylococcus aureus proteins Sbi and Efb recruit human plasmin to degrade complement C3 and C3b. PLoS One 7, e47638 (2012).
Acknowledgements
We thank W. Schneider, Medical University of Vienna, Austria, for advice; T. Hallström, Y. Lin, and S. Hälbich, Leibniz Institute for Natural Product Research and Infection Biology, Jena, for performing complement assays; W. Wilfert, Institute of Laboratory Medicine, University of Munich for lipid analyses; S. Schmidt, Institute for Experimental Neurology, Jena, for advice; and N. Buresch, Center for Neuropathology and Prion Research, Munich, for technical assistance; M. Jucker, Hertie Institute for Clinical Brain Research, University of Tübingen, for experimental support. This work was funded by the Deutsche Forschungsgemeinschaft (DFG): YI 133/2-1 to C.Y.; HA 1083/15-4 to A.J.R.H.; MO 3054/1-1 to S.M.; The German Collaborative Research Center (CRC124/2-C4), SK46/2 to C.S.; CRC124/2-C6 and DFG TR 1992 to P.F.Z.; the German Centre for Cardiovascular Research (DZHK MHA VD1.2), DFG SFB 1123/A1 and Z3, the European Research Council (ERC AdG 692511) to C.W.; SFB1123/Z01, INST409/150-1 FUGG to R.T.A.M.; Chinese National Natural Science Foundation (31770983) to D.H.; S.A. and L.D.H. are doctoral researchers at the International Leibniz Research School (ILRS) in Jena.
Author information
Authors and Affiliations
Contributions
These authors contributed equally: C.W., P.F.Z., C.S., and A.J.R.H.; C.Y. and A.J.R.H. designed and performed experiments and wrote the manuscript; S.A., P.F.Z., C.S. designed and performed experiments and contributed to writing the manuscript; C.W. provided critical intellectual input for experimental design and wrote the manuscript; A.B. provided the C5 siRNA; Z.M., S.K.M., C.Z., Y.L., S.N., M.W., L.P., D.H., S.V.B., P.S., M.B., R.T.A.M., S.S., M.H., L.D.H., H.-H.E., J.P., J.H., S.R., T.A., L.H., and C.J.B. performed experiments or analyzed the data.
Corresponding authors
Ethics declarations
Competing interests
C.Y., A.J.R.H., A.B., S.K.M., S.A., P.F.Z., and C.S. declare competing financial interests. C.Y., S.K.M., and A.J.R.H. are owners of Easemedcontrol R & D GmbH & Co KG Munich, Germany; A.B. is employed by Alnylam Pharmaceuticals Cambridge; Cambridge, MA, USA; C.Y. and A.J.R.H. have been named inventors on a pending patent application related to treatment and diagnosis of unresolvable inflammatory diseases (EP18183584.4); A.B. has been named as an inventor on patent applications related to C5 including PCT publication WO2014160129, and applications and patents based thereon; S.A., P.F.Z., and C.S. have been named inventors on a pending patent application (DE 10 2018 100 377.3).
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
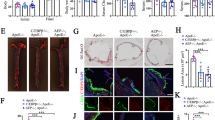
Extended Data Fig. 1 Lipid deposits, BBB, and ChP gene signatures.
a, Vacuole (Va) represents lipid. Intercellular lipid (green) between two epithelial cells was quantified. 68 intercellular spaces from 3 ApoE−/− and 67 intercellular spaces from 3 WT mice were analyzed. Bar represents 1 µm. b, Lipid in ApoE−/− ChPs by TEM. Lymphocytes (left panel); macrophages/dendritic cells (DC) (middle panel); and ependymal cells contain lipid (right panel). Vacuole (Va) represents lipid. Bar 1 µm. c, ChPs were stained for cytokeratin (keratin, red) and leukocytes (CD45, green) (left panel); collagen IV (Co-IV, green) and CD68 (red) (middle panel). TEM shows single macrophage-foam cell/DCs adjacent to microvilli. Bar 10 µm. d, ChPs were stained with Ig (red) as described in Methods. Bars 10 µm. e, PFA-perfused brains were stained for Ig (Ig, red) and blood vessels (Col-IV, green) in the cerebellum. Perivascular Ig adjacent to blood vessels was quantified as described in Methods. WT (n = 3 mice); ApoE−/− (n = 3); ND ApoE3 (n = 3); HFD ApoE3 (n = 3); ND ApoE4 (n = 3); HFD ApoE4 (n = 3). Bar 10 µm. f, Laser capture microdissection (LCM)-based expression microarrays of ChPs. Heatmaps show transcript levels in GO terms immune system process, transcription factor binding, cell junction, and ATP binding. g, Genes that were down-regulated in ApoE−/− CPs and rescued either in ApoE3-KI and in ND or HFD ApoE4-KI mice. WT (n = 5 mice); ApoE−/− (n = 4); ND ApoE3 (n = 6); HFD ApoE3 (n = 6); ND ApoE4 (n = 6); HFD ApoE4 (n = 6). Data in c,d are representative images from at least 3 biologically independent mouse samples. Data in a,e,g represent means ± s.e.m. Two-tailed Student´s t test was applied to a,e,g. Gene names and statistics in supplementary Tables 1, 3.
Extended Data Fig. 2 Complement constituents in mouse ChPs.
a, ChPs were stained for C1q (red) and C4 (green). Bar 100 μm. b, C5 siRNA treatment blocks C5 protein deposition in ApoE−/− ChPs. c, ChPs were stained for C3. Ig represents lipid. d, Serum C3 and C5. Serum C3 and C5 protein levels were measured by ELISA. ApoE−/−(n = 6 mice), HFD ApoE4 (n = 5). e, High resolution confocal microscopy shows colocalization of ApoE4 (ApoE, red) and Ig (green, represents lipid) in HFD ApoE4-KI ChPs. ApoE−/− ChPs serve as negative controls for ApoE staining. f, Complement regulators are expressed in ChPs. WT (n = 5 mice); ApoE−/−(n = 4); ND ApoE3 (n = 6); HFD ApoE3 (n = 6); ND ApoE4 (n = 6); HFD ApoE4 (n = 6). g, ChP Factor H expressed between WT and ApoE−/− mice. WT (n = 5); ApoE−/−(n = 4.) h, ChP factor H protein in ChPs. White arrows indicate lipid positive areas. Data in a,b,c,e,h are representative images from at least 3 biologically independent mouse samples. Data in d,f,g represent means ± s.e.m. Two-tailed Student´s t test was applied to d,g; one-way ANOVA with Tukey posttest was applied to f; Gene names in supplementary Tabl. 3.
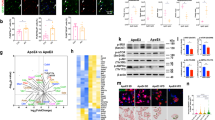
Extended Data Fig. 3 ApoE does not inhibit cleavage of C2 or C4 by C1s.
a, C1q binds immobilized malondialdehyde-modified LDL (MDA-LDL) and oxLDL but not native LDL or gelatin. b, ApoE isoforms in normal human serum (NHS) were added to MDA-LDL-coated microtiter plates and C4b deposition was determined by specific antisera. c,d IgM, MDA-LDL, and Aβ fibrils but not soluble Aβ activate complement and cause C3b deposition. BSA, gelatin as negative controls; e,f ApoE3 was incubated with either (e) C2 or (f) C4 in the presence of C1s. C2 and C4 were cleaved to their active forms C2a (α´30) and C4b (α´83) via C1s as revealed by the cleavage products in western blot analyses. g, ApoE3 has no cofactor activity for factor I in the cleavage of C4b to inactive iC4b. ApoE3 was incubated together with factor I, C4BP and C4b, and cleavage products were detected by western blot analysis as indicated (α´25 and α´13). Full scanned blot images in e,f,g are available from source data figures. Data in a-d represent means ± s.e.m. of three independent experiments. Two-tailed Student’s t test. Data in e,f,g are representative from 3 independent experiments.
Extended Data Fig. 4 ApoE binds to C1q but not to other complement components.
a, ApoE isoforms bind to the C1 complex, but not to C4 or C2. Biotinylated ApoE was immobilized on streptavidin-coated sensors and incubated with C1 complex, C4, C2, or buffer. b, The C1 complex binds to immobilized ApoE isoforms. c, ApoE isoforms bind to C1 and factor H, but not to C3 or C3b. d, NHS-derived C1 binds to immobilized plasma-purified ApoE3 and to recombinant ApoE isoforms. e, C1q binds to immobilized plasma-purified ApoE3 and to all recombinant ApoE isoforms. f, Plasma-purified C1q was coated on a sensor chip (CM5) and plasma-derived ApoE (62-1000 nM) was injected into the fluid phase (75 mM NaCl, 5 mM HEPES, 1 mM CaCl2). g, Mannose-binding lectin (MBL) does not bind to C1q as determined by biolayer interferometry. h, Apolipoprotein A (ApoA) does not bind to C1q as determined by biolayer interferometry. i, C1q-ApoE complexes revealed by proximity ligation assay (PLA) on cultured human apoptotic cells (THP-1) were detectable when treated with NHS but not with C1q-depleted serum (dNHS). Data represent mean fluorescence intensity (MFI) ± s.e.m. of 16 cells for each group. Bar 10 μm. Data in b,c,d,e represent means ± s.e.m. of at least three independent experiments. Data a,f,g and h represent means of at least two independent experiments. Two-tailed Student’s t test.
Extended Data Fig. 5 ApoE binds to the activated C1q; LDLR and C1sC1r tetramers do not compete with C1q-ApoE binding.
a, ApoE-C1q interaction is dependent on Ca2+. Real-time binding of ApoE to C1q was followed using biosensor analyses. Binding of ApoE to C1q is reduced in a dose-dependent manner upon increasing amounts of EGTA (0.1–3 mM). b,c, co-immunoprecipitation of C1q-ApoE complexes; (b) anti C1q antiserum precipitate C1q-ApoE complexes composed of purified proteins with activated C1q, but not with inactive C1q from NHS; (c) Anti-ApoE antiserum precipitates C1q-ApoE complexes but no complexes from NHS. C1q-ApoE complexes were eluted with glycine buffer, then, C1q or ApoE proteins were separated by SDS-PAGE and immunoblotted using goat anti-C1q antiserum (left panel of b, and c) or goat anti ApoE antiserum (right panel of b) separately. Full scanned blot images in b,c are available from source data figures. d, ApoE peptide P139-152 but not ApoE peptide P30-40 competes with immobilized ApoE3 for binding to C1q in a dose-dependent manner. e, C1q antibody binding to C1q is not affected by SDS. f, C1q and LDLR bind simultaneously to ApoE. 20 nM C1q was incubated with increasing concentrations of LDLR to immobilized ApoE and binding of C1q and LDLR was followed by ELISA. Background binding of anti C1q and anti LDLR antisera to immobilized ApoE were set as 0%. g, ApoE does not compete with C1sC1r tetramers for binding to C1q. C1q in addition to increasing amounts of C1sC1r tetramers was added to immobilized ApoE3 and C1q binding was determined. Data in d-g represent means ± s.e.m. of at least three independent experiments. Two-tailed Student´s t test. Data in a,b,c are representatives of 3 independent experiments.
Extended Data Fig. 6 Complement constituents in human ChPs and AD plaques.
a-b, Human ChP sections were stained for C1q (green) / C3 (red) (a) and C1q (green)/ApoE (red) (b). c,d, ChP sections were stained for CD68+ macrophages/DCs (c) and collagen IV (Col-IV) to mark basement membranes. Phase contrast shows lipid deposits in ChPs. e, ChP sections were stained for ApoE (green) and factor H (red); no primary antibody as control (NA). f,g, human brain sections were stained for Aβ (green) / ApoE (red) (left panels), Tau phosphorylation (pTau, green) / ApoE (red) (middle panels), and C1q (green) / ApoE (red) (right panels) (f). Blue for nuclei. No primary antibody as control (g). h, AD brain parenchyma sections were stained for C3 (red) / ApoE (green). Bar 100 μm for a-h; Data in a-h are representative images from at least 3 biologically independent samples.
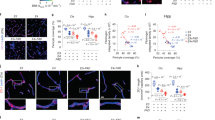
Extended Data Fig. 7 Complement constituents in mouse brain.
a, 16-week old APPPS1-21 mouse brain sections were stained with Aβ/ApoE complexes (red) by PLA, methoxy X04 for Aβ plaque (blue). High resolution confocal images show the spatial location of Aβ-ApoE complexes and Aβ plaque in 3D view (lower panel). Bars represent 10 µm. b, Brain sections were stained with methoxy-X04, ApoE, and LAMP1; the size of areas covered by methoxy-X04, ApoE, and LAMP1 was determined. ApoE/X04 and LAMP1/X04 (X04 > 150 µm2) were quantified. n = 123 plaques from 4 control mice, 147 plaques from 5 treated mice. Bars 100 µm. c, Aβ plaque was stained with methoxy X04 (X04). Number of plaques per section and number of plaque per area were quantified. control (n = 4 mice), C5 (n = 5). Bar 1000 µm. d, Total plaque volume was determined in 3D, plaques were further grouped according to the plaque volume. n = 71 random fields from 4 control mice, 88 fields from 5 C5 siRNA treated mice. Bar 100 µm. e, 8-week old C57BL6 brain cortex sections were examined for the presence of C1q-ApoE complexes with methoxy X04. ApoE, or C1q only antisera were used as negative controls. Bar represents 10 µm. Data in a,e are representative images from at least 3 biologically independent mouse samples. Data in b,c,d represent means ± s.e.m. Two-tailed Student´s t test was applied to b,c,d; Two-way ANOVA was applied to c,d.
Extended Data Fig. 8 Complement and atherosclerosis.
a, Expression microarray analyses of aortas. Heatmaps show GO terms leukocyte migration, complement activation, phagocytosis, and cellular response to lipid. 6 weeks WT (n = 3 mice); 32 weeks WT (n = 3); 6 weeks ApoE−/− (n = 3); 32 weeks ApoE−/− (n = 3). b, aorta alternative complement pathway genes (factor B, factor H, factor D) mRNA expression in 6 weeks and 32 weeks old WT and ApoE−/− mouse aortas. 6 weeks WT (n = 3 mice); 32 weeks WT (n = 3); 6 weeks ApoE−/− (n = 3); 32 weeks ApoE−/− (n = 3). c,d, plasma cholesterol and body weight. e,f blood leukocytes and percentage. For c-f, control (n =11 mice); C5 siRNA (n =12 mice). g, blood CD4+ T cells, CD8+ T cells, and B220+ B cells by flow cytometry. Control (n = 6 mice); C5 siRNA (n = 6 mice). h, super-resolution microscopy shows colocalization of C1q (green) and ApoE (red) in human atherosclerotic plaque. Representative images from at least 3 biologically independent mouse samples. Bar 5 μm. Data in b,c,d,e,f,g represent means ± s.e.m. Two-tailed Student´s t test was applied to c.d.e.f.g; one-way ANOVA with Tukey posttest was applied to b; abbreviations: WBC, white blood cells; RBC, red blood cells; PLT, platelets; LYM, lymphocytes; MO, monocytes; GRA, granulocytes. Gene names and statistics in supplementary Table 7.
Extended Data Fig. 9 Graphical presentation of the body of in vivo data.
a, Pleiotropic impacts of single ApoE or single C1q molecules in brain as reported by others. Microglia cells are the major source of brain C1q. In response to Aβ plaques, resting microglia cells differentiate into plaque-associated microglia cells. Single actions of ApoE and C1q have been reported to be involved in multiple pathways as indicated in the Figure. Inactive C1q (yellow), activated C1q (red). b, Graphical presentation of the body of in vivo data. Three types of unresolvable inflammatory conditions were studied in 7 mouse models and in translational studies of human tissues, that is choroid plexus, aorta, and brain parenchyma.
Supplementary information
Supplementary Tables
Supplementary Tables 1–8
Supplementary Video 1
Aβ plaque and C1q-ApoE complex in 3D.
Supplementary Video 2
Aβ plaque and Aβ-ApoE complex in 3D.
Source data
Source data Extended Data Fig. 3
Unprocessed Western Blots gels for extended figure 3.
Source data Extended Data Fig. 5
Unprocessed Western Blots gels for extended figure 5.
Rights and permissions
About this article
Cite this article
Yin, C., Ackermann, S., Ma, Z. et al. ApoE attenuates unresolvable inflammation by complex formation with activated C1q. Nat Med 25, 496–506 (2019). https://doi.org/10.1038/s41591-018-0336-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-018-0336-8
This article is cited by
-
Targeting synapse function and loss for treatment of neurodegenerative diseases
Nature Reviews Drug Discovery (2024)
-
Canonical and non-canonical roles of complement in atherosclerosis
Nature Reviews Cardiology (2024)
-
Kupffer cells dictate hepatic responses to the atherogenic dyslipidemic insult
Nature Cardiovascular Research (2024)
-
How does apolipoprotein E genotype influence the relationship between physical activity and Alzheimer’s disease risk? A novel integrative model
Alzheimer's Research & Therapy (2023)
-
Analysis of complement system and its related factors in Alzheimer’s disease
BMC Neurology (2023)