Abstract
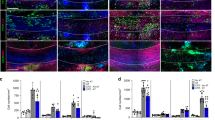
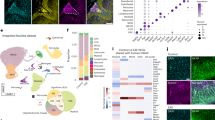
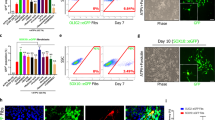
Multiple sclerosis (MS) is characterized by an immune system attack targeting myelin, which is produced by oligodendrocytes (OLs). We performed single-cell transcriptomic analysis of OL lineage cells from the spinal cord of mice induced with experimental autoimmune encephalomyelitis (EAE), which mimics several aspects of MS. We found unique OLs and OL precursor cells (OPCs) in EAE and uncovered several genes specifically alternatively spliced in these cells. Surprisingly, EAE-specific OL lineage populations expressed genes involved in antigen processing and presentation via major histocompatibility complex class I and II (MHC-I and -II), and in immunoprotection, suggesting alternative functions of these cells in a disease context. Importantly, we found that disease-specific oligodendroglia are also present in human MS brains and that a substantial number of genes known to be susceptibility genes for MS, so far mainly associated with immune cells, are expressed in the OL lineage cells. Finally, we demonstrate that OPCs can phagocytose and that MHC-II-expressing OPCs can activate memory and effector CD4-positive T cells. Our results suggest that OLs and OPCs are not passive targets but instead active immunomodulators in MS. The disease-specific OL lineage cells, for which we identify several biomarkers, may represent novel direct targets for immunomodulatory therapeutic approaches in MS.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
A web resource for browsing differential gene expression data for the single-cell data can be accessed at https://ki.se/en/mbb/oligointernode. Raw data are deposited in GEO, accession number GSE113973.
References
Patsopoulos, N.et al. The Multiple Sclerosis Genomic Map: Role of peripheral immune cells and resident microglia in susceptibility. Preprint at bioRxiv https://doi.org/10.1101/143933 (2017)
Skene, N. G. & Grant, S. G. Identification of vulnerable cell types in major brain disorders using single cell transcriptomes and expression weighted cell type enrichment. Front. Neurosci. 10, 16 (2016).
Marques, S. et al. Transcriptional convergence of oligodendrocyte lineage progenitors during development. Dev. Cell 46, 504–517.e7 (2018).
Marques, S. et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–1329 (2016).
Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013).
Klinghoffer, R. A., Hamilton, T. G., Hoch, R. & Soriano, P. An allelic series at the PDGFalphaR locus indicates unequal contributions of distinct signaling pathways during development. Dev. Cell 2, 103–113 (2002).
Kang, S. H., Fukaya, M., Yang, J. K., Rothstein, J. D. & Bergles, D. E. NG2+CNS glial progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron 68, 668–681 (2010).
Emery, B. et al. Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell 138, 172–185 (2009).
Ogata, T. et al. Hes1 functions downstream of growth factors to maintain oligodendrocyte lineage cells in the early progenitor stage. Neuroscience 176, 132–141 (2011).
Tripathi, R. B., Rivers, L. E., Young, K. M., Jamen, F. & Richardson, W. D. NG2 glia generate new oligodendrocytes but few astrocytes in a murine experimental autoimmune encephalomyelitis model of demyelinating disease. J. Neurosci. 30, 16383–16390 (2010).
Zawadzka, M. et al. CNS-resident glial progenitor/stem cells produce Schwann cells as well as oligodendrocytes during repair of CNS demyelination. Cell Stem Cell 6, 578–590 (2010).
Gregory, A. P. et al. TNF receptor 1 genetic risk mirrors outcome of anti-TNF therapy in multiple sclerosis. Nature 488, 508–511 (2012).
Capello, E., Voskuhl, R. R., McFarland, H. F. & Raine, C. S. Multiple sclerosis: re-expression of a developmental gene in chronic lesions correlates with remyelination. Ann. Neurol. 41, 797–805 (1997).
Enevold, C. et al. Multiple sclerosis and polymorphisms of innate pattern recognition receptors TLR1-10, NOD1-2, DDX58, and IFIH1. J. Neuroimmunol. 212, 125–131 (2009).
Haile, Y. et al. Granzyme B-inhibitor serpina3n induces neuroprotection in vitro and in vivo. J. Neuroinflamm. 12, 157 (2015).
Lee, S. C. & Raine, C. S. Multiple sclerosis: oligodendrocytes in active lesions do not express class II major histocompatibility complex molecules. J. Neuroimmunol. 25, 261–266 (1989).
Sibinga, N. E., Feinberg, M. W., Yang, H., Werner, F. & Jain, M. K. Macrophage-restricted and interferon gamma-inducible expression of the allograft inflammatory factor-1 gene requires Pu.1. J. Biol. Chem. 277, 16202–16210 (2002).
Lin, W., Harding, H. P., Ron, D. & Popko, B. Endoplasmic reticulum stress modulates the response of myelinating oligodendrocytes to the immune cytokine interferon-gamma. J. Cell Biol. 169, 603–612 (2005).
Kobayashi, K. S. & van den Elsen, P. J. NLRC5: a key regulator of MHC class I-dependent immune responses. Nat. Rev. Immunol. 12, 813–820 (2012).
Bergsteindottir, K., Brennan, A., Jessen, K. R. & Mirsky, R. In the presence of dexamethasone, gamma interferon induces rat oligodendrocytes to express major histocompatibility complex class II molecules. Proc. Natl Acad. Sci. USA 89, 9054–9058 (1992).
Brosius Lutz, A. et al. Schwann cells use TAM receptor-mediated phagocytosis in addition to autophagy to clear myelin in a mouse model of nerve injury. Proc. Natl Acad. Sci. USA 114, E8072–E8080 (2017).
Bettelli, E. et al. Myelin oligodendrocyte glycoprotein-specific T cell receptor transgenic mice develop spontaneous autoimmune optic neuritis. J. Exp. Med. 197, 1073–1081 (2003).
Zeis, T., Enz, L. & Schaeren-Wiemers, N. The immunomodulatory oligodendrocyte. Brain Res. 1641, 139–148 (2016).
Peferoen, L., Kipp, M., van der Valk, P., van Noort, J. M. & Amor, S. Oligodendrocyte-microglia cross-talk in the central nervous system. Immunology 141, 302–313 (2014).
Zeis, T. & Schaeren-Wiemers, N. Lame ducks or fierce creatures? The role of oligodendrocytes in multiple sclerosis. J. Mol. Neurosci. 35, 91–100 (2008).
Moyon, S. et al. Demyelination causes adult CNS progenitors to revert to an immature state and express immune cues that support their migration. J. Neurosci. 35, 4–20 (2015).
Huynh, J. L. et al. Epigenome-wide differences in pathology-free regions of multiple sclerosis-affected brains. Nat. Neurosci. 17, 121–130 (2014).
Traka, M., Podojil, J. R., McCarthy, D. P., Miller, S. D. & Popko, B. Oligodendrocyte death results in immune-mediated CNS demyelination. Nat. Neurosci. 19, 65–74 (2016).
Matsuoka, T. et al. Neural crest origins of the neck and shoulder. Nature 436, 347–355 (2005).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–13 (2011).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Norton, W. T. & Poduslo, S. E. Myelination in rat brain: method of myelin isolation. J. Neurochem. 21, 749–757 (1973).
Larocca, J. N. & Norton, W. T. Isolation of myelin. Curr. Protoc. Cell Biol. 33, 3.25.1–3.25.19 (2007).
Zeisel, A., Yitzhaky, A., Bossel Ben-Moshe, N. & Domany, E. An accessible database for mouse and human whole transcriptome qPCR primers. Bioinformatics 29, 1355–1356 (2013).
Angerer, P. et al. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics 32, 1241–1243 (2016).
Bivand, R., Hauke, J. & Kossowski, T. Computing the Jacobian in Gaussian spatial autoregressive models: an illustrated comparison of available methods. Geogr. Anal. 45, 150–179 (2013).
Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290.e17 (2017).
Finak, G. et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Killcoyne, S., Carter, G. W., Smith, J. & Boyle, J. in Protein Networks and Pathway Analysis (eds. Nikolsky, Y. & Bryant, J.) 219–239 (Humana Press, New York, 2009).
Bindea, G. et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093 (2009).
Wolfert, M. A. & Boons, G. J. Adaptive immune activation: glycosylation does matter. Nat. Chem. Biol. 9, 776–784 (2013).
Huang, Y. & Sanguinetti, G. BRIE: transcriptome-wide splicing quantification in single cells. Genome Biol. 18, 123 (2017).
Thorvaldsdóttir, H., Robinson, J. T. & Mesirov, J. P. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013).
Durinck, S., Spellman, P. T., Birney, E. & Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191 (2009).
Quinlan, A. R. BEDTools: the Swiss-army tool for genome feature analysis. Curr. Protoc. Bioinformatics 47, 11.12.1–11.12.34 (2014).
Acknowledgements
We would like to thank A. Nanni, A. Moshref, Tony Jimenez-Beristain and J. Söderlund for laboratory management and support. We thank Eukaryotic Single Cell Genomics Facility, WABI Long Term Bioinformatic Support (Leif Wigge) at SciLifeLab, the FACS facilities at CMB (B. Panagel), Science for Life Laboratory, the National Genomics Infrastructure and Uppmax for providing assistance in massive parallel sequencing and computational infrastructure. We wish to acknowledge A. G. Rothfuchs for advice and reagents, M. Jagodic for advice, M. Bartosovic for assistance, R. Berglund and M. N’diaye for providing the tools to perform the phagocytosis experiments. The bioinformatics computations were performed on resources provided by the Swedish National Infrastructure for Computing at UPPMAX, Uppsala University. Postmortem MS tissue used for IHC was provided via a UK prospective donor scheme with full ethical approval from the UK Multiple Sclerosis Tissue Bank (MREC/02/02/39). D.P.V. would like to acknowledge the University of Sydney HPC service for providing HPC resources that have contributed to the research reported in this paper. This work was supported in part by a University of Sydney HPC Grand Challenge Award. D.P.V. was supported in part by a Boehringer Ingelheim Travel Grant. C.f.-C. is funded by a Wellcome Trust Investigator award. A.W. is funded by UK Multiple Sclerosis Society. S.J. is funded by European Union, Horizon 2020, Marie-Skłodowska Curie Actions (grant no. EC 789492); A.M.F. by the European Committee for Treatment and Research of Multiple Sclerosis (ECTRIMS). E.A. is funded by European Union, Horizon 2020, Marie-Skłodowska Curie Actions (grant no. SOLO 794689). Work in G.C.-B.’s research group was supported by Swedish Research Council (grant no. 2015-03558), European Union (Horizon 2020 Research and Innovation Programme/European Research Council Consolidator Grant EPIScOPE, grant agreement 681893), Swedish Brain Foundation (grant no. FO2017-0075), Ming Wai Lau Centre for Reparative Medicine, Petrus och Augusta Hedlunds Foundation (grant nos. M-2014-0041 and M-2016-0428) and Karolinska Institutet.
Author information
Authors and Affiliations
Contributions
A.M.F., D.V.B and G.C.-B. conceived the project, designed the study and interpreted results. A.M.F., S.M. and A.O.G-C. performed EAE model and A.M.F and S.M. collected single cells to generate single-cell sequencing data. D.V.B and E.A. performed computational analyses. A.M.F. and M.M. designed, performed and analyzed most in vitro OPC experiments and S. performed the phagocytosis experiments together with A.M.F. A.O.G-C. and A.M.F. designed, performed and analyzed all experiments involving co-cultures with immune cells. S.J., A.W. and C.f.-C. provided the human postmortem MS brain tissue and performed the human IHC analysis. D.P.V. provided support for computational analysis. E.M.F. provided RNAscope ISH expertise and performed all videos. A.M.F, D.V.B and G.C.-B. wrote the manuscript with feedback from all authors.
Corresponding authors
Ethics declarations
Competing interests
The G.C.-B., A.W. and C.f.-C. research groups have received funding from F. Hoffmann–La Roche, Ltd. for other research projects in this area.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9
Supplementary Video 1
MOL2 cells express MHC-II genes
Supplementary Video 2
OPCs express MHC-II genes
Supplementary Video 3
OL lineage cells express MHC-II genes
Supplementary Video 4
OL lineage cells express MHC-II genes
Supplementary Video 5
OL lineage cells express MHC-II genes and few Aif1 molecules
Supplementary Video 6
OL lineage cells express MHC-II genes
Supplementary Video 7
Microglia processes touch OL lineage cells
Supplementary Video 8
MHC-II positive cells surround OL lineage cells in human MS patient samples
Supplementary Video 9
OL lineage cells from human MS patient samples express MHC-II genes
Supplementary Table 1
Differential gene expression and gene module association
Supplementary Table 2
Splicing events and exon inclusion or exclusion events
Supplementary Table 3
Differential gene expression between CTRL and EAE in MOLs and OPCs
Supplementary Table 4
Non-MHC MS susceptibility genes expressed in microglia and OPCs in EAE and CTRL and significant differential expression of these genes across the major level 1 cluster
Rights and permissions
About this article
Cite this article
Falcão, A.M., van Bruggen, D., Marques, S. et al. Disease-specific oligodendrocyte lineage cells arise in multiple sclerosis. Nat Med 24, 1837–1844 (2018). https://doi.org/10.1038/s41591-018-0236-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-018-0236-y
This article is cited by
-
Single-nucleus transcriptomic analysis reveals the relationship between gene expression in oligodendrocyte lineage and major depressive disorder
Journal of Translational Medicine (2024)
-
Microglia-derived exosomes modulate myelin regeneration via miR-615-5p/MYRF axis
Journal of Neuroinflammation (2024)
-
Glial-restricted progenitor cells: a cure for diseased brain?
Biological Research (2024)
-
Myeloid cell replacement is neuroprotective in chronic experimental autoimmune encephalomyelitis
Nature Neuroscience (2024)
-
hnRNP A1 dysfunction alters RNA splicing and drives neurodegeneration in multiple sclerosis (MS)
Nature Communications (2024)