Abstract
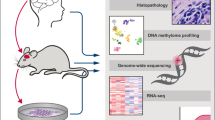
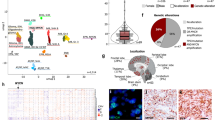
Brain tumors are the leading cause of cancer-related death in children. Genomic studies have provided insights into molecular subgroups and oncogenic drivers of pediatric brain tumors that may lead to novel therapeutic strategies. To evaluate new treatments, better preclinical models adequately reflecting the biological heterogeneity are needed. Through the Children’s Oncology Group ACNS02B3 study, we have generated and comprehensively characterized 30 patient-derived orthotopic xenograft models and seven cell lines representing 14 molecular subgroups of pediatric brain tumors. Patient-derived orthotopic xenograft models were found to be representative of the human tumors they were derived from in terms of histology, immunohistochemistry, gene expression, DNA methylation, copy number, and mutational profiles. In vivo drug sensitivity of targeted therapeutics was associated with distinct molecular tumor subgroups and specific genetic alterations. These models and their molecular characterization provide an unprecedented resource for the cancer community to study key oncogenic drivers and to evaluate novel treatment strategies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
PDOX lines described here, as well as additional lines being developed and characterized, are catalogued online and shared with other researchers at the Brain Tumor Resource Laboratory (http://www.btrl.org). In addition, an online summary of each model and interactive access to the molecular and histopathological data can be found in the R2 PDX Explorer (http://www.r2platform.com/pdxexplorer). Short-read sequencing data are available at the European Genome-phenome Archive (http://www.ebi.ac.uk/ega/), hosted by the European Bioinformatics Institute, under accession EGAS00001002536. Methylation and gene expression data have been deposited in the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) under accessions GSE99994 and GSE99961.
References
Taylor, M. D. et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 123, 465–472 (2012).
Johann, P. D. et al. Atypical teratoid/rhabdoid tumors are comprised of three epigenetic subgroups with distinct enhancer landscapes. Cancer Cell 29, 379–393 (2016).
Torchia, J. et al. Integrated (epi)-genomic analyses identify subgroup-specific therapeutic targets in CNS rhabdoid tumors. Cancer Cell 30, 891–908 (2016).
Sturm, D. et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22, 425–437 (2012).
Johnson, R. A. et al. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature 466, 632–636 (2010).
Witt, H. et al. Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell 20, 143–157 (2011).
Gajjar, A., Pfister, S. M., Taylor, M. D. & Gilbertson, R. J. Molecular insights into pediatric brain tumors have the potential to transform therapy. Clin. Cancer Res. 20, 5630–5640 (2014).
Capper, D. et al. DNA methylation-based classification of central nervous system tumours. Nature 555, 469–474 (2018).
Louis, D. N. et al. The2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 131, 803–820 (2016).
Pajtler, K. W. et al. The current consensus on the clinical management of intracranial ependymoma and its distinct molecular variants. Acta Neuropathol. 133, 5–12 (2017).
Pajtler, K. W. et al. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell 27, 728–743 (2015).
Shou, Y. et al. A five-gene hedgehog signature developed as a patient preselection tool for hedgehog inhibitor therapy in medulloblastoma. Clin. Cancer Res. 21, 585–593 (2015).
Gilbertson, R. J. Brain tumors provide new clues to the source of cancer stem cells: does oncology recapitulate ontogeny? Cell Cycle 5, 135–137 (2006).
Packer, R. J. et al. Phase III study of craniospinal radiation therapy followed by adjuvant chemotherapy for newly diagnosed average-risk medulloblastoma. J Clin. Oncol. 24, 4202–4208 (2006).
Gao, H. et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 21, 1318–1325 (2015).
Townsend, E. C. et al. The public repository of xenografts enables discovery and randomized phase II-like trials in mice. Cancer Cell 30, 183 (2016).
Bruna, A. et al. A biobank of breast cancer explants with preserved intra-tumor heterogeneity to screen anticancer compounds. Cell 167, 260–274.e222 (2016).
Houghton, P. J. et al. The pediatric preclinical testing program: description of models and early testing results. Pediatr. Blood Cancer 49, 928–940 (2007).
Zhao, X. et al. Global gene expression profiling confirms the molecular fidelity of primary tumor-based orthotopic xenograft mouse models of medulloblastoma. Neuro. Oncol. 14, 574–583 (2012).
Grasso, C. S. et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat. Med. 21, 827 (2015).
Goldenberg, D. M. & Pavia, R. A. In vivo horizontal oncogenesis by a human tumor in nude mice. Proc. Natl Acad. Sci. USA 79, 2389–2392 (1982).
Triviai, I. et al. Endogenous retrovirus induces leukemia in a xenograft mouse model for primary myelofibrosis. Proc.Natl Acad. Sci. USA 111, 8595–8600 (2014).
Hovestadt, V. et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature 510, 537–541 (2014).
Sturm, D. et al. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell 164, 1060–1072 (2016).
Korshunov, A. et al. H3-/IDH-wild type pediatric glioblastoma is comprised of molecularly and prognostically distinct subtypes with associated oncogenic drivers. Acta Neuropathol. 134, 507–516 (2017).
Mackay, A. et al. Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell 32, 520–537 (2017). e525.
Northcott, P. A. et al. The whole-genome landscape of medulloblastoma subtypes. Nature 547, 311–317 (2017).
Callari, M. et al. Computational approach to discriminate human and mouse sequences in patient-derived tumour xenografts. BMC Genomics 19, 19 (2018).
Northcott, P. A. et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 511, 428–434 (2014).
Pajtler, K. W. et al. Molecular heterogeneity and CXorf67 alterations in posterior fossa group A (PFA) ependymomas. Acta Neuropathol. 136, 211–226 (2018).
Kilday, J. P. et al. Copy number gain of 1q25 predicts poor progression-free survival for pediatric intracranial ependymomas and enables patient risk stratification: a prospective European clinical trial cohort analysis on behalf of the Children’s Cancer Leukaemia Group (CCLG), Societe Francaise d’Oncologie Pediatrique (SFOP), and International Society for Pediatric Oncology (SIOP). Clin. Cancer Res. 18, 2001–2011 (2012).
Parker, M. et al. C11orf95-RELA fusions drive oncogenic NF-kappaB signalling in ependymoma. Nature 506, 451–455 (2014).
Biegel, J. A. Molecular genetics of atypical teratoid/rhabdoid tumor. Neurosurg. Focus 20, E11 (2006).
Shih, D. J. et al. Cytogenetic prognostication within medulloblastoma subgroups. J. Clin. Oncol. 32, 886–896 (2014).
Pei, Y. et al. HDAC and PI3K antagonists cooperate to inhibit growth of MYC-driven medulloblastoma. Cancer Cell 29, 311–323 (2016).
Lee, S. J. et al. Sonic hedgehog-induced histone deacetylase activation is required for cerebellar granule precursor hyperplasia in medulloblastoma. PLoS ONE. 8, e71455 (2013).
Spiller, S. E., Ditzler, S. H., Pullar, B. J. & Olson, J. M. Response of preclinical medulloblastoma models to combination therapy with 13-cis retinoic acid and suberoylanilide hydroxamic acid (SAHA). J. Neurooncol. 87, 133–141 (2008).
Cook Sangar, M. L. et al. Inhibition of CDK4/6 by palbociclib significantly extends survival in medulloblastoma patient-derived xenograft mouse models. Clin. Cancer Res. 23, 5802–5813 (2017).
Varley, K. E. et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 23, 555–567 (2013).
Goodspeed, A., Heiser, L. M., Gray, J. W. & Costello, J. C. Tumor-derived cell lines as molecular models of cancer pharmacogenomics. Mol. Cancer Res. 14, 3–13 (2016).
Ramaswamy, V. et al. Medulloblastoma subgroup-specific outcomes in irradiated children: who are the true high-risk patients? Neuro Oncol. 18, 291–297 (2016).
Hidalgo, M. et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 4, 998–1013 (2014).
Peereboom, D. M. et al. Phase II trial of erlotinib with temozolomide and radiation in patients with newly diagnosed glioblastoma multiforme. J. Neurooncol. 98, 93–99 (2010).
Wen, P. Y. et al. Phase I/II study of erlotinib and temsirolimus for patients with recurrent malignant gliomas: North American Brain Tumor Consortium trial 04-02. Neuro Oncol. 16, 567–578 (2014).
Raizer, J. J. et al. A phase II study of bevacizumab and erlotinib after radiation and temozolomide in MGMT unmethylated GBM patients. J. Neurooncol. 126, 185–192 (2016).
Sflomos, G. et al. A preclinical model for ERα-positive breast cancer points to the epithelial microenvironment as determinant of luminal phenotype and hormone response. Cancer Cell 29, 407–422 (2016).
Stewart, E. et al. Orthotopic patient-derived xenografts of paediatric solid tumours. Nature 549, 96–100 (2017).
Eirew, P. et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 518, 422–426 (2015).
Hill, R. M. et al. Combined MYC and P53 defects emerge at medulloblastoma relapse and define rapidly progressive, therapeutically targetable disease. Cancer Cell 27, 72–84 (2015).
Lee, J. et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 9, 391–403 (2006).
Saeed, A. I. et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34, 374–378 (2003).
Smyth, G. K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3 (2004).
Wilson, C. L. & Miller, C. J. Simpleaffy: a bioconductor package for affymetrix quality control and data analysis. Bioinformatics 21, 3683–3685 (2005).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009).
Hovestadt, V. et al. Robust molecular subgrouping and copy-number profiling of medulloblastoma from small amounts of archival tumour material using high-density DNA methylation arrays. Acta Neuropathol. 125, 913–916 (2013).
van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Mazor, T. et al. DNA methylation and somatic mutations converge on the cell cycle and define similar evolutionary histories in brain tumors. Cancer Cell 28, 307–317 (2015).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Futreal, P. A. et al. A census of human cancer genes. Nat. Rev. Cancer 4, 177–183 (2004).
Zhao, M., Sun, J. & Zhao, Z. TSGene: a web resource for tumor suppressor genes. Nucleic Acids Res. 41, D970–D976 (2013).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
McPherson, A. et al. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput. Biol. 7, e1001138 (2011).
Campbell, I. Chi-squared and Fisher-Irwin tests of two-by-two tables with small sample recommendations. Stat. Med. 26, 3661–3675 (2007).
Richardson, J. T. The analysis of 2×2 contingency tables—yet again. Stat. Med. 30, 890; author reply 891–892. (2011).
Acknowledgements
We are grateful to the patients and families who consented to provide tissue to generate these resources. We thank A. Richards, M. Merrill, D. Acorn, M. Biery, A. Wittmann, and L. Sieber for experimental contributions. We thank the DKFZ Genomics and Proteomics Core Facility, DKFZ Heidelberg, Germany, and the AMC Department of Oncogenomics, Amsterdam, The Netherlands, for performing high-throughput sequencing and microarray analyses to a very high standard. We also thank the DKFZ data management group, especially I. Scholz, for their excellent support in processing the sequencing data, and Z. Gu for the defuse pipeline. We thank X.-N. Li, R. J. Wechsler-Reya, and T. Milde for allowing us to list their PDOX lines on our websites. This work was supported by NIH 1U10CA180886-01 (J.M.O.), NIH 1R01CA155360 (J.M.O.), NIH R01 CA114567 (J.M.O.), the Seattle Run of Hope, the Seattle Children’s Brain Tumor Research Endowment, the Dutch Cancer Foundations KWF (2010-4713) (M.K.) and KIKA (90) (M.K.), Deutsche Krebshilfe (111537) (S.M.P., M.K.), BMBF (01KT1605) (S.M.P., M.K.), IMI-JU ITCC-P4 (116064) (D.T.W.J., J.K., S.M.P., M.K.), and the Helmholtz International Graduate School for Cancer Research (S.B.).
Author information
Authors and Affiliations
Contributions
S.B., S.E.S.L., S.N.G., M.W.N., H.S.-C, E.J.G., B.C., A.D.S., K.L.B., N.L.M., F.P., B.S., A.K., K.D.P., J.D., S.H., and S.D. performed and/or coordinated the experimental work. S.B., S.E.S.L., S.N.G., M.W.N., H.S.-C., E.J.G, B.C., A.D.S., K.L.B., V.H., A.K., G.P.B., P.A.N., K.D.P., S.H., S.D., P.L., L.C., D.T.W.J., J.K., and M.K. performed data analysis and interpretation. S.B., S.E.S.L., S.N.G., S.M.P., M.K., and J.M.O. provided project leadership. S.B., S.E.S.L., S.N.G., S.M.P., M.K., and J.M.O. prepared the initial manuscript and figures. All authors contributed to the critical review of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10
Supplementary Table 1
PDOX model information
Supplementary Table 2
Pathological characterization of PDOX models and matching human tumors
Supplementary Table 3
Overview about molecular data
Supplementary Table 4
Molecular alterations in PDOX models and matching human tumors
Supplementary Table 5
Genes with increased and decreased expression in PDOX compared to human tumors
Supplementary Table 6
Primers used for Sanger sequencing validations and for qRT–PCR
Rights and permissions
About this article
Cite this article
Brabetz, S., Leary, S.E.S., Gröbner, S.N. et al. A biobank of patient-derived pediatric brain tumor models. Nat Med 24, 1752–1761 (2018). https://doi.org/10.1038/s41591-018-0207-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-018-0207-3
This article is cited by
-
Marinopyrrole derivative MP1 as a novel anti-cancer agent in group 3 MYC-amplified Medulloblastoma
Journal of Experimental & Clinical Cancer Research (2024)
-
Generation, evolution, interfering factors, applications, and challenges of patient-derived xenograft models in immunodeficient mice
Cancer Cell International (2023)
-
Spatial transcriptomic analysis of Sonic hedgehog medulloblastoma identifies that the loss of heterogeneity and promotion of differentiation underlies the response to CDK4/6 inhibition
Genome Medicine (2023)
-
Molecular and functional profiling of chemotolerant cells unveils nucleoside metabolism-dependent vulnerabilities in medulloblastoma
Acta Neuropathologica Communications (2023)
-
Metabolism-based targeting of MYC via MPC-SOD2 axis-mediated oxidation promotes cellular differentiation in group 3 medulloblastoma
Nature Communications (2023)