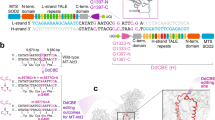
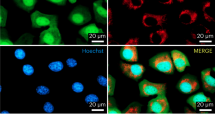
Abstract
Mutations of the mitochondrial genome (mtDNA) underlie a substantial portion of mitochondrial disease burden. These disorders are currently incurable and effectively untreatable, with heterogeneous penetrance, presentation and prognosis. To address the lack of effective treatment for these disorders, we exploited a recently developed mouse model that recapitulates common molecular features of heteroplasmic mtDNA disease in cardiac tissue: the m.5024C>T tRNAAla mouse. Through application of a programmable nuclease therapy approach, using systemically administered, mitochondrially targeted zinc-finger nucleases (mtZFN) delivered by adeno-associated virus, we induced specific elimination of mutant mtDNA across the heart, coupled to a reversion of molecular and biochemical phenotypes. These findings constitute proof of principle that mtDNA heteroplasmy correction using programmable nucleases could provide a therapeutic route for heteroplasmic mitochondrial diseases of diverse genetic origin.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
All next-generation sequencing data generated in the present study are available from the BioProject database using accession PRJNA479953. All other datasets and materials are available from the corresponding authors upon reasonable request.
References
Gorman, G. S. et al. Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Ann. Neurol. 77, 753–759 (2015).
Wachsmuth, M., Hubner, A., Li, M., Madea, B. & Stoneking, M. Age-related and heteroplasmy-related variation in human mtDNA copy number. PLoS Genet. 12, e1005939 (2016).
Gorman, G. S. et al. Mitochondrial diseases. Nat. Rev. Dis. Primers 2, 16080 (2016).
Bacman, S. R., Williams, S. L., Pinto, M., Peralta, S. & Moraes, C. T. Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs. Nat. Med. 19, 1111–1113 (2013).
Gammage, P. A., Rorbach, J., Vincent, A. I., Rebar, E. J. & Minczuk, M. Mitochondrially targeted ZFNs for selective degradation of pathogenic mitochondrial genomes bearing large-scale deletions or point mutations. EMBO Mol. Med. 6, 458–466 (2014).
Reddy, P. et al. Selective elimination of mitochondrial mutations in the germline by genome editing. Cell 161, 459–469 (2015).
Gammage, P. A., Moraes, C. T. & Minczuk, M. Mitochondrial genome engineering: the revolution may not be CRISPR-Ized. Trends Genet. 34, 101–110 (2018).
Alexeyev, M., Shokolenko, I., Wilson, G. & LeDoux, S. The maintenance of mitochondrial DNA integrity—critical analysis and update. Cold Spring Harb. Perspect. Biol. 5, a012641 (2013).
Peeva, V. et al. Linear mitochondrial DNA is rapidly degraded by components of the replication machinery. Nat. Commun. 9, 1727 (2018).
Minczuk, M., Papworth, M. A., Kolasinska, P., Murphy, M. P. & Klug, A. Sequence-specific modification of mitochondrial DNA using a chimeric zinc finger methylase. Proc. Natl Acad. Sci. USA 103, 19689–19694 (2006).
Minczuk, M., Kolasinska-Zwierz, P., Murphy, M. P. & Papworth, M. A. Construction and testing of engineered zinc-finger proteins for sequence-specific modification of mtDNA. Nat. Protoc. 5, 342–356 (2010).
Minczuk, M., Papworth, M. A., Miller, J. C., Murphy, M. P. & Klug, A. Development of a single-chain, quasi-dimeric zinc-finger nuclease for the selective degradation of mutated human mitochondrial DNA. Nucleic Acids Res. 36, 3926–3938 (2008).
Gammage, P. A. et al. Near-complete elimination of mutant mtDNA by iterative or dynamic dose-controlled treatment with mtZFNs. Nucleic Acids Res. 44, 7804–7816 (2016).
Gaude, E. et al. NADH shuttling couples cytosolic reductive carboxylation of glutamine with glycolysis in cells with mitochondrial dysfunction. Mol. Cell 69, 581–593.e7 (2018).
Kauppila, J. H. et al. A phenotype-driven approach to generate mouse models with pathogenic mtDNA mutations causing mitochondrial disease. Cell Rep. 16, 2980–2990 (2016).
Gammage, P. A., Van Haute, L. & Minczuk, M. Engineered mtZFNs for manipulation of human mitochondrial DNA heteroplasmy. Methods Mol. Biol. 1351, 145–162 (2016).
Pulicherla, N. et al. Engineering liver-detargeted AAV9 vectors for cardiac and musculoskeletal gene transfer. Mol. Ther. 19, 1070–1078 (2011).
Yarham, J. W., Elson, J. L., Blakely, E. L., McFarland, R. & Taylor, R. W. Mitochondrial tRNA mutations and disease. Wiley Interdiscip. Rev. RNA 1, 304–324 (2010).
Jazayeri, M. et al. Inducible expression of a dominant negative DNA polymerase-γ depletes mitochondrial DNA and produces a rho0 phenotype. J. Biol. Chem. 278, 9823–9830 (2003).
Birsoy, K. et al. An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 162, 540–551 (2015).
Holt, I. J., Harding, A. E. & Morgan-Hughes, J. A. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature 331, 717–719 (1988).
Wallace, D. C. et al. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science 242, 1427–1430 (1988).
Craven, L. et al. Pronuclear transfer in human embryos to prevent transmission of mitochondrial DNA disease. Nature 465, 82–85 (2010).
Tachibana, M. et al. Towards germline gene therapy of inherited mitochondrial diseases. Nature 493, 627–631 (2013).
Floros, V. I. et al. Segregation of mitochondrial DNA heteroplasmy through a developmental genetic bottleneck in human embryos. Nat. Cell Biol. 20, 144–151 (2018).
Yamada, M. et al. Genetic drift can compromise mitochondrial replacement by nuclear transfer in human oocytes. Cell Stem Cell 18, 749–754 (2016).
Vafai, S. B. & Mootha, V. K. Mitochondrial disorders as windows into an ancient organelle. Nature 491, 374–383 (2012).
Viscomi, C., Bottani, E. & Zeviani, M. Emerging concepts in the therapy of mitochondrial disease. Biochim. Biophys. Acta 1847, 544–557 (2015).
Pfeffer, G. et al. New treatments for mitochondrial disease-no time to drop our standards. Nat. Rev. Neurol. 9, 474–481 (2013).
Bacman, S. R. et al. MitoTALEN reduces mutant mtDNA load and restores tRNAAla levels in a mouse model of heteroplasmic mtDNA mutation. Nat. Med. https://doi.org/10.1038/s41591-018-0166-8 (2018).
Beilstein, K., Wittmann, A., Grez, M. & Suess, B. Conditional control of mammalian gene expression by tetracycline-dependent hammerhead ribozymes. ACS Synth. Biol. 4, 526–534 (2015).
Pearce, S. F. et al. Maturation of selected human mitochondrial tRNAs requires deadenylation. eLife 6, e27596 (2017).
Mackay, G. M., Zheng, L., van den Broek, N. J. & Gottlieb, E. Analysis of cell metabolism using LC–MS and isotope tracers. Methods Enzymol. 561, 171–196 (2015).
Acknowledgements
This work was supported by the Medical Research Council (MC_U105697135 and MC_UU_00015/4 to M.M., MC_UU_12022/6 to C.F. and MC_UU_00015/5 to M.Z.), ERC Advanced Grant (FP7-322424 to M.Z.), NRJ-Institut de France (to M.Z.) and the Max Planck Society (to J.B.S.). P.R.-G. was supported by ‘Fundação para a Ciência e a Tecnologia’ (PD/BD/105750/2014). We acknowledge the important contribution to model development made by N.-G. Larsson, which was essential to this work. We are grateful to the personnel at Phenomics Animal Care Facility, Cambridge, UK, for their technical support in managing our mouse colonies. We are grateful to M. Rice, Phenomics Animal Care Facility, for technical assistance with viral administration. We thank R. Dirksen (MPI, Cologne, Germany) for isolation and immortalization of the MEFs. All FACS experiments were performed at the NIHR BRC Cell Phenotyping Hub, Cambridge, UK, by C. Bowman, E. Perez, J. Markovic Djuric and A. Petrunkina-Harrison.
Author information
Authors and Affiliations
Contributions
P.A.G. designed the research, performed biochemical, in vitro and in vivo experiments, analyzed the data and wrote the paper. C.V. performed the in vivo experiments. M.-L.S. contributed to model characterization. A.S.H.C. and E.G. performed the mass spectrometry-based metabolomic experiments and analyzed the data. C.A.P. and L.V.H. performed biochemical experiments and analyzed the data. B.J.M. performed biochemical and immunofluorescence experiments. P.R.-G. and R.C. performed the histological experiments. L.Z. designed and assembled the ZFP library. E.J.R. oversaw the ZFP library preparation. M.Z. oversaw the in vivo experiments. C.F. oversaw the mass spectrometry-based metabolomic experiments. J.B.S. provided cell and mouse models and contributed to model characterization. M.M. oversaw the project and co-wrote the paper, with all authors’ involvement.
Corresponding authors
Ethics declarations
Competing interests
E.J.R. and L.Z. are full-time employees of Sangamo Therapeutics.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 and Supplementary Tables 1 and 2
Rights and permissions
About this article
Cite this article
Gammage, P.A., Viscomi, C., Simard, ML. et al. Genome editing in mitochondria corrects a pathogenic mtDNA mutation in vivo. Nat Med 24, 1691–1695 (2018). https://doi.org/10.1038/s41591-018-0165-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-018-0165-9
This article is cited by
-
Strand-selective base editing of human mitochondrial DNA using mitoBEs
Nature Biotechnology (2024)
-
Teamwork makes the dream work: functional collaborations between families, scientists, and healthcare providers to drive progress in the treatment of Leigh Syndrome
Orphanet Journal of Rare Diseases (2023)
-
Mitochondrial heterogeneity in diseases
Signal Transduction and Targeted Therapy (2023)
-
Efficient elimination of MELAS-associated m.3243G mutant mitochondrial DNA by an engineered mitoARCUS nuclease
Nature Metabolism (2023)
-
Trends and prospects in mitochondrial genome editing
Experimental & Molecular Medicine (2023)