Abstract
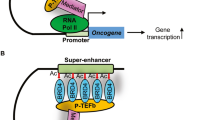
Cancer cells rely on dysregulated gene expression. This establishes specific transcriptional addictions that may be therapeutically exploited. Yet, the mechanisms that are ultimately responsible for these addictions are poorly understood. Here, we investigated the transcriptional dependencies of transformed cells to the transcription factors YAP and TAZ. YAP/TAZ physically engage the general coactivator bromodomain-containing protein 4 (BRD4), dictating the genome-wide association of BRD4 to chromatin. YAP/TAZ flag a large set of enhancers with super-enhancer-like functional properties. YAP/TAZ-bound enhancers mediate the recruitment of BRD4 and RNA polymerase II at YAP/TAZ-regulated promoters, boosting the expression of a host of growth-regulating genes. Treatment with small-molecule inhibitors of BRD4 blunts YAP/TAZ pro-tumorigenic activity in several cell or tissue contexts, causes the regression of pre-established, YAP/TAZ-addicted neoplastic lesions and reverts drug resistance. This work sheds light on essential mediators, mechanisms and genome-wide regulatory elements that are responsible for transcriptional addiction in cancer and lays the groundwork for a rational use of BET inhibitors according to YAP/TAZ biology.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The RNA-seq and ChIP–seq data generated in this study have been deposited in the GEO database under accession GSE102409. The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Bradner, J. E., Hnisz, D. & Young, R. A. Transcriptional addiction in cancer. Cell 168, 629–643 (2017).
Villicana, C., Cruz, G. & Zurita, M. The basal transcription machinery as a target for cancer therapy. Cancer Cell Int. 14, 18 (2014).
Andrieu, G., Belkina, A. C. & Denis, G. V. Clinical trials for BET inhibitors run ahead of the science. Drug Discov. Today Technol. 19, 45–50 (2016).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
Shi, J. & Vakoc, C. R. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol. Cell 54, 728–736 (2014).
Stathis, A. et al. Clinical response of carcinomas harboring the BRD4-NUT oncoprotein to the targeted bromodomain inhibitor OTX015/MK-8628. Cancer Discov. 6, 492–500 (2016).
Loven, J. et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013).
Pott, S. & Lieb, J. D. What are super-enhancers? Nat. Genet. 47, 8–12 (2015).
Azzolin, L. et al. YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell 158, 157–170 (2014).
Bai, H. et al. Yes-associated protein regulates the hepatic response after bile duct ligation. Hepatology 56, 1097–1107 (2012).
Cai, J. et al. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 24, 2383–2388 (2010).
Chen, Q. et al. A temporal requirement for Hippo signaling in mammary gland differentiation, growth, and tumorigenesis. Genes Dev. 28, 432–437 (2014).
Zanconato, F. et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 17, 1218–1227 (2015).
Su, T. et al. Two-signal requirement for growth-promoting function of Yap in hepatocytes. eLife 4, e02948 (2015).
Zhang, W. et al. Downstream of mutant KRAS, the transcription regulator YAP is essential for neoplastic progression to pancreatic ductal adenocarcinoma. Sci. Signal. 7, ra42 (2014).
Taniguchi, K. et al. A gp130–Src–YAP module links inflammation to epithelial regeneration. Nature 519, 57–62 (2015).
Zanconato, F., Cordenonsi, M. & Piccolo, S. YAP/TAZ at the roots of cancer. Cancer Cell 29, 783–803 (2016).
Panciera, T. et al. Induction of expandable tissue-specific stem/progenitor cells through transient expression of YAP/TAZ. Cell Stem Cell 19, 725–737 (2016).
Johnson, R. & Halder, G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat. Rev. Drug Discov. 13, 63–79 (2014).
Rafiee, M. R., Girardot, C., Sigismondo, G. & Krijgsveld, J. Expanding the circuitry of pluripotency by selective isolation of chromatin-associated proteins. Mol. Cell 64, 624–635 (2016).
Skibinski, A. et al. The Hippo transducer TAZ interacts with the SWI/SNF complex to regulate breast epithelial lineage commitment. Cell Rep. 6, 1059–1072 (2014).
Oh, H. et al. Yorkie promotes transcription by recruiting a histone methyltransferase complex. Cell Rep. 8, 449–459 (2014).
Stein, C. et al. YAP1 exerts its transcriptional control via TEAD-mediated activation of enhancers. PLoS Genet. 11, e1005465 (2015).
Chen, H. I. & Sudol, M. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc. Natl Acad. Sci. USA 92, 7819–7823 (1995).
Shu, S. et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 529, 413–417 (2016).
Wang, Y. et al. CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell 163, 174–186 (2015).
Galli, G. G. et al. YAP drives growth by controlling transcriptional pause release from dynamic enhancers. Mol. Cell 60, 328–337 (2015).
Devaiah, B. N. et al. BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat. Struct. Mol. Biol. 23, 540–548 (2016).
Tropberger, P. et al. Regulation of transcription through acetylation of H3K122 on the lateral surface of the histone octamer. Cell 152, 859–872 (2013).
Zhan, T., Rindtorff, N. & Boutros, M. Wnt signaling in cancer. Oncogene 36, 1461–1473 (2017).
Swellam, M. et al. Aberrant methylation of APC and RARβ2 genes in breast cancer patients. IUBMB Life 67, 61–68 (2015).
Bernasconi, E. et al. Preclinical evaluation of the BET bromodomain inhibitor BAY 1238097 for the treatment of lymphoma. Br. J. Haematol. 178, 936–948 (2017).
Camargo, F. D. et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 17, 2054–2060 (2007).
Dong, J. et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133 (2007).
Yimlamai, D. et al. Hippo pathway activity influences liver cell fate. Cell 157, 1324–1338 (2014).
Storz, P. Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nat. Rev. Gastroenterol. Hepatol. 14, 296–304 (2017).
Kim, M. H. et al. Actin remodeling confers BRAF inhibitor resistance to melanoma cells through YAP/TAZ activation. EMBO J. 35, 462–478 (2016).
Lin, L. et al. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat. Genet. 47, 250–256 (2015).
Zuo, Q. et al. AXL/AKT axis mediated-resistance to BRAF inhibitor depends on PTEN status in melanoma. Oncogene 37, 3275–3289 (2018).
Lee, B. S. et al. Hippo effector YAP directly regulates the expression of PD-L1 transcripts in EGFR-TKI-resistant lung adenocarcinoma. Biochem. Biophys. Res. Commun. 491, 493–499 (2017).
Enzo, E. et al. Aerobic glycolysis tunes YAP/TAZ transcriptional activity. EMBO J. 34, 1349–1370 (2015).
Roessler, S. et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 70, 10202–10212 (2010).
Rizos, H. et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: spectrum and clinical impact. Clin. Cancer Res. 20, 1965–1977 (2014).
Xu, Y. & Vakoc, C. R. Targeting cancer cells with BET bromodomain inhibitors. Cold Spring Harb. Perspect. Med. 7, a026674 (2017).
Cordenonsi, M. et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 147, 759–772 (2011).
Aragona, M. et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154, 1047–1059 (2013).
Bisgrove, D. A., Mahmoudi, T., Henklein, P. & Verdin, E. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc. Natl Acad. Sci. USA 104, 13690–13695 (2007).
Hotta, A. et al. Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nat. Methods 6, 370–376 (2009).
Garcia-Gutierrez, P., Mundi, M. & Garcia-Dominguez, M. Association of bromodomain BET proteins with chromatin requires dimerization through the conserved motif B. J. Cell Sci. 125, 3671–3680 (2012).
Azzolin, L. et al. Role of TAZ as mediator of Wnt signaling. Cell 151, 1443–1456 (2012).
Dupont, S. et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011).
Martello, G. et al. A microRNA targeting Dicer for metastasis control. Cell 141, 1195–1207 (2010).
Hughes, C. S. et al. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 10, 757 (2014).
Means, A. L. et al. Pancreatic epithelial plasticity mediated by acinar cell transdifferentiation and generation of nestin-positive intermediates. Development 132, 3767–3776 (2005).
Zhang, N. et al. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev. Cell 19, 27–38 (2010).
Sansom, O. J. et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 18, 1385–1390 (2004).
Schuler, M., Dierich, A., Chambon, P. & Metzger, D. Efficient temporally controlled targeted somatic mutagenesis in hepatocytes of the mouse. Genesis 39, 167–172 (2004).
Wagner, K. U. et al. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 25, 4323–4330 (1997).
Belteki, G. et al. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res. 33, e51 (2005).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Karolchik, D. et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–D496 (2004).
Jin, F. et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503, 290–294 (2013).
Chapuy, B. et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell 24, 777–790 (2013).
Zhang, Y. et al. Model-based analysis of ChIP–seq (MACS). Genome Biol. 9, R137 (2008).
Picaud, S. et al. RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain. Proc. Natl Acad. Sci. USA 110, 19754–19759 (2013).
Mori, M. et al. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell 156, 893–906 (2014).
Fontanals-Cirera, B. et al. Harnessing BET inhibitor sensitivity reveals AMIGO2 as a melanoma survival gene. Mol. Cell 68, 731–744.e9 (2017).
Roessler, S. et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology 142, 957–966.e12 (2012).
Du, P., Kibbe, W. A. & Lin, S. M. lumi: a pipeline for processing Illumina microarray. Bioinformatics 24, 1547–1548 (2008).
Adorno, M. et al. A mutant-p53/Smad complex opposes p63 to empower TGFβ-induced metastasis. Cell 137, 87–98 (2009).
Acknowledgements
We are grateful to J. C. Marine (Leuven Center for Cancer Biology), J. Kim (KAIST) for the gift of the cell lines; D. J. Pan (University of Texas), A. R. Clarke (Cardiff University), F. Camargo (Boston Children’s Hospital) and P. Chambon (University of Strasbourg) for the gifts of mice. MMTV-cre mice were purchased from The Jackson Laboratory, where they were kindly deposited by L. Hennighausen. This work is supported by the AIRC Special Program Molecular Clinical Oncology ‘5 per mille’, by an AIRC PI-Grant to S.P. and by the Epigenetics Flagship project CNR-MIUR. This project has received funding from the European Research Council under the European Union’s Horizon 2020 research and innovation programme (DENOVOSTEM grant agreement no. 670126).
Author information
Authors and Affiliations
Contributions
F.Z., M.C. and S.P. designed the study, analyzed the data and wrote the manuscript. F.Z., G.B. and L.F. performed the experiments. M.Forcato and S.B. performed the bioinformatics analysis. L.A., E.Q., D.D.B., V.G. and M.Fassan performed the animal experiments and the histological analysis. P.L. and B.H. provided the reagents and advice for the animal experiments. G.S. and J.K. performed the mass spectrometry. A.M. performed the initial experiments of this study.
Corresponding authors
Ethics declarations
Competing interests
B.H. and P.L. are employees of Bayer AG. S.P. is a consultant for and received institutional grants from Bayer AG.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7
Supplementary Table 1
YAP/TAZ nuclear interactors
Supplementary Table 2
Direct YAP/TAZ target genes inhibited by JQ1
Rights and permissions
About this article
Cite this article
Zanconato, F., Battilana, G., Forcato, M. et al. Transcriptional addiction in cancer cells is mediated by YAP/TAZ through BRD4. Nat Med 24, 1599–1610 (2018). https://doi.org/10.1038/s41591-018-0158-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-018-0158-8
This article is cited by
-
A phenotypic screening approach to target p60AmotL2-expressing invasive cancer cells
Journal of Experimental & Clinical Cancer Research (2024)
-
BET inhibitors drive Natural Killer activation in non-small cell lung cancer via BRD4 and SMAD3
Nature Communications (2024)
-
Nuclear phosphoinositide signaling promotes YAP/TAZ-TEAD transcriptional activity in breast cancer
The EMBO Journal (2024)
-
A chaperone-like function of FUS ensures TAZ condensate dynamics and transcriptional activation
Nature Cell Biology (2024)
-
Targeting ONECUT3 blocks glycolytic metabolism and potentiates anti-PD-1 therapy in pancreatic cancer
Cellular Oncology (2024)