Abstract
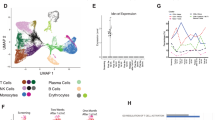
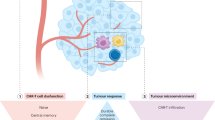
We identified genetic mutations in CD19 and loss of heterozygosity at the time of CD19– relapse to chimeric antigen receptor (CAR) therapy. The mutations are present in the vast majority of resistant tumor cells and are predicted to lead to a truncated protein with a nonfunctional or absent transmembrane domain and consequently to a loss of surface antigen. This irreversible loss of CD19 advocates for an alternative targeting or combination CAR approach.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Grupp, S. A. et al. Blood 122, 67 (2013).
Maude, S. L. et al. N. Engl. J. Med. 378, 439–448 (2018).
Maude, S. L. et al. Blood 128, 2801 (2016).
Buechner, J. et al. EHA 17, abstr. S476 (2017).
Wang, Z., Wu, Z., Liu, Y. & Han, W. J. Hematol. Oncol. 10, 53 (2017).
Ruella, M. & Maus, M. V. Comput. Struct. Biotechnol. J. 14, 357–362 (2016).
Riester, M. et al. Source Code Biol. Med. 11, 13 (2016).
Sotillo, E. et al. Cancer Discov. 5, 1282–1295 (2015).
Shen, S. et al. Proc. Natl. Acad. Sci. USA 111, E5593–E5601 (2014).
Jacoby, E. et al. Nat. Commun. 7, 12320 (2016).
Gardner, R. et al. Blood 127, 2406–2410 (2016).
Sommermeyer, D. et al. Leukemia 31, 2191–2199 (2017).
Shah, N. N. et al. Blood 128, 650 (2016).
Topp, M. S. et al. J. Clin. Oncol. 32, 4134–4140 (2014).
Zugmaier, G. et al. Blood 126, 2578–2584 (2015).
Cibulskis, K. et al. Nat. Biotechnol. 31, 213–219 (2013).
Ye, K., Schulz, M. H., Long, Q., Apweiler, R. & Ning, Z. Bioinformatics 25, 2865–2871 (2009).
Sherry, S. T. et al. Nucleic Acids Res. 29, 308–311 (2001).
Forbes, S. A. et al. Nucleic Acids Res. 43, D805–D811 (2015).
Li, H. & Durbin, R. Bioinformatics 25, 1754–1760 (2009).
McKenna, A. et al. Genome Res. 20, 1297–1303 (2010).
DePristo, M. A. et al. Nat. Genet. 43, 491–498 (2011).
Acknowledgements
The authors thank the patients and their families for participating in these clinical trials and A. Abrams for figure design.
Author information
Authors and Affiliations
Contributions
J.E.L., M.Q., S.A.G., M.B., B.D.M., E.R.N., H. Bittencourt, H.H., J.B., S.M.D., and M.V. contributed patient samples for sequencing. P.A.W. provided clinical data on the patients. R.J.L. oversaw the DNA- and RNA-seq. E.J.O. and M.R. analyzed and interpreted the sequencing data. X.H., P.P., and K.N. analyzed and interpreted the flow cytometry MRD assay data. W.W., J.A.E., H. Bitter, M.M., J.L.B., C.T., and S.P. provided guidance and scientific input. E.J.O. wrote the paper, with contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
The research samples used in this manuscript come from patients treated on clinical trials conducted by Novartis. J.E.L., M.Q., H. Bittencourt, S.M.D, and S.A.G. consult for Novartis. E.J.O., X.H., C.T., P.A.W., R.J.L., M.R., J.L.B., H.Bitter, M.M., P.P., S.P., J.A.E., and W.W. are employed by Novartis or were employed by Novartis at the time of manuscript preparation.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 and Supplementary Tables 1–6
Supplementary Data Table
Supplementary Data Table 1
Rights and permissions
About this article
Cite this article
Orlando, E.J., Han, X., Tribouley, C. et al. Genetic mechanisms of target antigen loss in CAR19 therapy of acute lymphoblastic leukemia. Nat Med 24, 1504–1506 (2018). https://doi.org/10.1038/s41591-018-0146-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-018-0146-z
This article is cited by
-
Challenges and strategies associated with CAR-T cell therapy in blood malignancies
Experimental Hematology & Oncology (2024)
-
Immune evasion in cell-based immunotherapy: unraveling challenges and novel strategies
Journal of Biomedical Science (2024)
-
Alternative splicing and related RNA binding proteins in human health and disease
Signal Transduction and Targeted Therapy (2024)
-
CAR T cell therapy for patients with solid tumours: key lessons to learn and unlearn
Nature Reviews Clinical Oncology (2024)
-
Challenges in the discovery of tumor-specific alternative splicing-derived cell-surface antigens in glioma
Scientific Reports (2024)