Abstract
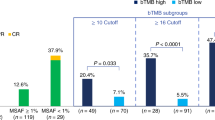
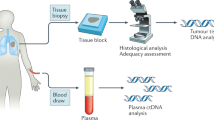
Although programmed death-ligand 1–programmed death 1 (PD-L1–PD-1) inhibitors are broadly efficacious, improved outcomes have been observed in patients with high PD-L1 expression or high tumor mutational burden (TMB). PD-L1 testing is required for checkpoint inhibitor monotherapy in front-line non-small-cell lung cancer (NSCLC). However, obtaining adequate tumor tissue for molecular testing in patients with advanced disease can be challenging. Thus, an unmet medical need exists for diagnostic approaches that do not require tissue to identify patients who may benefit from immunotherapy. Here, we describe a novel, technically robust, blood-based assay to measure TMB in plasma (bTMB) that is distinct from tissue-based approaches. Using a retrospective analysis of two large randomized trials as test and validation studies, we show that bTMB reproducibly identifies patients who derive clinically significant improvements in progression-free survival from atezolizumab (an anti-PD-L1) in second-line and higher NSCLC. Collectively, our data show that high bTMB is a clinically actionable biomarker for atezolizumab in NSCLC.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Rittmeyer, A. et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 389, 255–265 (2017).
Borghaei, H. et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N. Engl. J. Med. 373, 1627–1639 (2015).
Brahmer, J. et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N. Engl. J. Med. 373, 123–135 (2015).
Reck, M. et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N. Engl. J. Med. 375, 1823–1833 (2016).
Fehrenbacher, L. et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 387, 1837–1846 (2016).
Carbone, D. P. et al. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N. Engl. J. Med. 376, 2415–2426 (2017).
Garon, E. B. Cancer immunotherapy trials not immune from imprecise selection of patients. N. Engl. J. Med. 376, 2483–2485 (2017).
Hui, R. et al. Pembrolizumab as first-line therapy for patients with PD-L1-positive advanced non-small cell lung cancer: a phase 1 trial. Ann. Oncol. 28, 874–881 (2017).
Tischer, B., Kim, E., Peters, M. & Hirsh, V. Physician patterns of care in patients with EGFR mutation+ NSCLC: an international survey into testing and treatment choice. J. Thorac. Oncol. 12, S1199–S1200 (2017).
Lim, C. et al. Biomarker testing and time to treatment decision in patients with advanced nonsmall-cell lung cancer. Ann. Oncol. 26, 1415–1421 (2015).
Green, M. R. et al. Molecular testing prior to first-line therapy in patients with stage IV nonsquamous non-small cell lung cancer (NSCLC): a survey of U.S. medical oncologists. J. Clin. Oncol. 26, abstr. 8097 (2014).
Rizvi, N. A. et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015).
Rooney, M. S., Shukla, S. A., Wu, C. J., Getz, G. & Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61 (2015).
Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013).
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Kowanetz, M. et al. Tumor mutation burden (TMB) is associated with improved efficacy of atezolizumab in 1L and 2L+ NSCLC patients. J. Thorac. Oncol. 12, S321–S322 (2017).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Le, D. T. et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017).
Johnson, D. B. et al. Targeted next generation sequencing identifies markers of response to PD-1 blockade. Cancer Immunol. Res. 4, 959–967 (2016).
Chalmers, Z. R. et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 9, 34 (2017).
Campesato, L. F. et al. Comprehensive cancer-gene panels can be used to estimate mutational load and predict clinical benefit to PD-1 blockade in clinical practice. Oncotarget 6, 34221–34227 (2015).
Rosenberg, J. E. et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 387, 1909–1920 (2016).
Abbosh, C. et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545, 446–451 (2017).
Jamal-Hanjani, M. et al. Tracking the evolution of non-small-cell lung cancer. N. Engl. J. Med. 376, 2109–2121 (2017).
Wan, J. C. et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer 17, 223–238 (2017).
Siravegna, G., Marsoni, S., Siena, S. & Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 14, 531–548 (2017).
Chaudhuri, A. A. et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 7, 1394–1403 (2017).
Frampton, G. M. et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 31, 1023–1031 (2013).
Fabrizio, D. et al. Analytic validation of a next generation sequencing assay to identify tumor mutational burden from blood (bTMB) to support investigation of an anti-PD-L1 agent, atezolizumab, in a first line non-small cell lung cancer trial (BFAST). Ann. Oncol. 28, mdx363.018 (2017).
He, J. et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood 127, 3004–3014 (2016).
Friedman, J. H. & Roosen, C. B. An introduction to multivariate adaptive regression splines. Stat. Methods Med. Res. 4, 197–217 (1995).
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Chen, L., Liu, P., Evans, T. C. Jr & Ettwiller, L. M. DNA damage is a pervasive cause of sequencing errors, directly confounding variant identification. Science 355, 752–756 (2017).
Sun, J. X. et al. A computational method for somatic versus germline variant status determination from targeted next-generation sequencing of clinical cancer specimens without a matched normal control. Cancer Res. 74, 1893 (2014).
Sun, J. X. et al. A computational approach to distinguish somatic vs. germline origin of genomic alterations from deep sequencing of cancer specimens without a matched normal. PLoS Comput. Biol. 14, e1005965 (2018).
Diaz, L. A. Jr et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 486, 537–540 (2012).
Newman, A. M. et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 20, 548–554 (2014).
Mok, T. et al. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin. Cancer Res. 21, 3196–3203 (2015).
Acknowledgements
We thank the patients, their families and the clinical study-site investigators and staff for their contributions to the study. In addition, we acknowledge E. White, M. Coyne, T. Brennan, M. Kennedy, J. He, S. Zhong, G. Young, J. Ma, M. Zhao and G. Frampton for their contributions. This study was funded by F. Hoffmann-La Roche. Editorial support for this manuscript was provided by M. Balbas of Health Interactions and funded by F. Hoffmann-La Roche.
Author information
Authors and Affiliations
Contributions
D.R.G. interpreted the experiments and wrote the paper. S.M.P. designed and interpreted the experiments and wrote the paper. M.K. interpreted the experiments and wrote the paper. E.S. designed and interpreted the experiments and wrote the paper. W.Z. analyzed and interpreted the experiments and wrote the paper. Y.L. analyzed and interpreted the experiments and wrote the paper. A.R. designed and interpreted the experiments and wrote the paper. L.F. designed and interpreted the experiments and wrote the paper. G.O. designed, performed, analyzed and interpreted the experiments. C.M. analyzed the experiments. D.S.L. and D.L. performed and analyzed the experiments. J.S. performed and analyzed the experiments. L.A. interpreted the experiments and wrote the paper. T.R. interpreted the experiments and wrote the paper. C.A.C. designed, analyzed and interpreted the experiments and wrote the paper. P.S.H. interpreted the experiments and wrote the paper. A.S. designed and interpreted the experiments. M.B. designed and interpreted the experiments and wrote the paper. D.F. designed, performed, analyzed and interpreted the experiments and wrote the paper. T.M. interpreted the experiments and wrote the paper. D.S.S. designed, analyzed and interpreted the experiments and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare the following competing interests: D.R.G. has received a clinical trial grant and consultant fees from Genentech. Y.L., L.A., T.R., A.S. and M.B. are Genentech employees and holders of Roche stock. M.K., E.S., D.S.S., S.M.P., Y.L. and C.A.C. are Genentech employees and holders of Roche stock and have a patent pending for therapeutic and diagnostic methods for cancer. W.Z. is a Genentech employee and has received research funding from Roche. A.R. has received grants from AstraZeneca, Roche/Genentech, Bristol-Myers Squibb, Boehringer Ingelheim, Eli Lilly and Pfizer. L.F. declares no competing interests. D.F. is a Foundation Medicine employee and stockholder and has a patent pending for therapeutic and diagnostic methods for cancer. G.O., C.M., D.S.L., D.L. and J.S. are Foundation Medicine employees and stockholders. P.S.H. is a Genentech employee. T.M. has received grants/research support from AstraZeneca, Boehringer Ingelheim, Pfizer, Novartis, SFJ Pharmaceuticals, Roche, Merck Sharp & Dohme, Clovis Oncology, Bristol-Myers Squibb, Eisai and Taiho; speaker’s fees from AstraZeneca, Roche/Genentech, Pfizer, Eli Lilly, Bristol-Myers Squibb, Merck Sharp & Dohme, Novartis, Taiho and Ariad/Takeda; honoraria/honorarium from AstraZeneca, Roche/Genentech, Pfizer, Eli Lilly, Boehringer Ingelheim, Merck Serono, Merck Sharp & Dohme, Novartis, SFJ Pharmaceuticals, ACEA Biosciences, Vertex Pharmaceuticals, Bristol-Myers Squibb, OncoGenex Pharmaceuticals, Celgene and Ignyta; is a major stockholder in Sanomics and Cirina; participated in advisory boards for AstraZeneca, Roche/Genentech, Pfizer, Eli Lilly, Boehringer Ingelheim, Clovis Oncology, Merck Serono, Merck Sharp & Dohme, Novartis, SFJ Pharmaceuticals, ACEA Biosciences, Vertex Pharmaceuticals, Bristol-Myers Squibb, geneDecode, OncoGenex Technologies, Celgene, Ignyta and Cirina; and is on the board of directors for the International Association for the Study of Lung Cancer (IASLC), the Chinese Lung Cancer Research Foundation, the Chinese Society of Clinical Oncology (CSCO) and the Hong Kong Cancer Therapy Society (HKCTS).
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary Text and Figures
Supplementary Figures 1–11 and Supplementary Tables 1–7 and 9
Supplementary Table 8
Patient-level clinical variables, clinical outcome data and variant calls for bTMB in OAK and POPLAR
Rights and permissions
About this article
Cite this article
Gandara, D.R., Paul, S.M., Kowanetz, M. et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med 24, 1441–1448 (2018). https://doi.org/10.1038/s41591-018-0134-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-018-0134-3
This article is cited by
-
Blood-based biomarkers in patients with non-small cell lung cancer treated with immune checkpoint blockade
Journal of Experimental & Clinical Cancer Research (2024)
-
Targeting focal adhesion kinase boosts immune response in KRAS/LKB1 co-mutated lung adenocarcinoma via remodeling the tumor microenvironment
Experimental Hematology & Oncology (2024)
-
Tumor mutational burden predictability in head and neck squamous cell carcinoma patients treated with immunotherapy: systematic review and meta-analysis
Journal of Translational Medicine (2024)
-
Genomic profiling and clinical utility of circulating tumor DNA in metastatic prostate cancer: SCRUM-Japan MONSTAR SCREEN project
BJC Reports (2024)
-
Progress of immune checkpoint inhibitors therapy for non-small cell lung cancer with liver metastases
British Journal of Cancer (2024)