Abstract
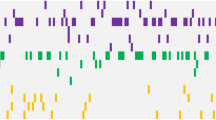
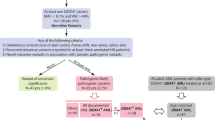
The pattern of somatic mutations observed at diagnosis of acute myeloid leukemia (AML) has been well-characterized. However, the premalignant mutational landscape of AML and its impact on risk and time to diagnosis is unknown. Here we identified 212 women from the Women’s Health Initiative who were healthy at study baseline, but eventually developed AML during follow-up (median time: 9.6 years). Deep sequencing was performed on peripheral blood DNA of these cases and compared to age-matched controls that did not develop AML. We discovered that mutations in IDH1, IDH2, TP53, DNMT3A, TET2 and spliceosome genes significantly increased the odds of developing AML. All subjects with TP53 mutations (n = 21 out of 21 patients) and IDH1 and IDH2 (n = 15 out of 15 patients) mutations eventually developed AML in our study. The presence of detectable mutations years before diagnosis suggests that there is a period of latency that precedes AML during which early detection, monitoring and interventional studies should be considered.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Mardis, E. R. et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 361, 1058–1066 (2009).
Ley, T. J. et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 363, 2424–2433 (2010).
Ding, L. et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481, 506–510 (2012).
Genovese, G. et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371, 2477–2487 (2014).
Jaiswal, S. et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371, 2488–2498 (2014).
Xie, M. et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 20, 1472–1478 (2014).
Coombs, C. C. et al. Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell 21, 374–382 (2017).
The Women’s Health Initiative Study Group. Design of the Women’s Health Initiative clinical trial and observational study. Control. Clin. Trials 19, 61–109 (1998).
Anderson, G. L. et al. Implementation of the Women’s Health Initiative study design. Ann. Epidemiol. 13, S5–S17 (2003).
Bergstralh, E. J., Kosanke, J. L.. & Jacobsen, S. J. Software for optimal matching in observational studies. Epidemiology 7, 331–332 (1996).
Bowman, R. L., Busque, L. & Levine, R. L. Clonal hematopoiesis and evolution to hematopoietic malignancies. Cell Stem Cell 22, 157–170 (2018).
Ward, P. S. et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17, 225–234 (2010).
Makishima, H. et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood 119, 3203–3210 (2012).
Zhang, S. J. et al. Genetic analysis of patients with leukemic transformation of myeloproliferative neoplasms shows recurrent SRSF2 mutations that are associated with adverse outcome. Blood 119, 4480–4485 (2012).
Olivier, M., Hollstein, M. & Hainaut, P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2, a001008 (2010).
Kadia, T. M. et al. TP53 mutations in newly diagnosed acute myeloid leukemia: clinicomolecular characteristics, response to therapy, and outcomes. Cancer 15, 3484–3491 (2016).
Samuelsen, S. O. A psudolikelihood approach to analysis of nested case–control studies. Biometrika 84, 379–394 (1997).
Palmisano, M. et al. NPM1 mutations are more stable than FLT3 mutations during the course of disease in patients with acute myeloid leukemia. Haematologica 92, 1268–1269 (2007).
Zink, F. et al. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood 130, 742–752 (2017).
Young, A. L., Challen, G. A., Birmann, B. M. & Druley, T. E. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat. Commun. 7, 12484 (2016).
Kim, S. S. et al. Loss-of-function mutations in the splicing factor ZRSR2 are common in blastic plasmacytoid dendritic cell neoplasm and have male predominance. Blood 122, 741 (2013).
Mori, T. et al. Somatic PHF6 mutations in1760 cases with various myeloid neoplasms. Leukemia 30, 2270–2273 (2016).
Lawrence, M. S. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014).
The Cancer Genome Atlas Research Network.. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013).
Papaemmanuil, E. et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 374, 2209–2221 (2016).
Enasidenib approved for AML, but best uses unclear. Cancer Discov. 7, OF4 (2017).
Lee, S. C. & Abdel-Wahab, O. Therapeutic targeting of splicing in cancer. Nat. Med. 22, 976–986 (2016).
Montalban-Bravo, G., Garcia-Manero, G. & Jabbour, E. Therapeutic choices after hypomethylating agent resistance for myelodysplastic syndromes. Curr. Opin. Hematol. 25, 146–153 (2018).
Sabapathy, K. & Lane, D. P. Therapeutic targeting of p53: all mutants are equal, but some mutants are more equal than others. Nat. Rev. Clin. Oncol. 15, 13–30 (2018).
Cimmino, L. et al. Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. Cell 170, 1079–1095 (2017).
Kircher, M., Sawyer, S. & Meyer, M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40, e3 (2012).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997 (2013).
Lai, Z. et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 44, e108 (2016).
Li, H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics 30, 2843–2851 (2014).
Acknowledgements
We acknowledge all our donors to Leukemia Fighters, without whom this work would not have been possible; the women who generously participated in the WHI study. We thank J. Z. Xiang and the Weill Cornell Genomics Core Facility as well as J. Catalano of the Englander Institute for Precision Medicine for assistance in sequencing; L.-B. Yan for technical assistance. The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, US Department of Health and Human Services through contracts N01WH22110, 24152, 32100-2, 32105-6, 32108-9, 32111-13, 32115, 32118-32119, 32122, 42107-26, 42129-32, and 44221, and the Cancer Center Support Grant NIH:NCI P30CA022453. We acknowledge the dedicated efforts of investigators and staff at the WHI clinical centers, the WHI Clinical Coordinating Center, and the National Heart, Lung and Blood program office (listing available at http://www.whi.org). We are additionally grateful for funding from the Sandra and Edward Meyer Cancer Center, which partially supported this study (D.C.H.).
Author information
Authors and Affiliations
Contributions
P.D., M.S.S., M.L.G., G.J.R. and D.C.H. designed and supervised the study; P.D., N.M.-T., M.L.G., G.J.R. and D.C.H. wrote the manuscript; P.D. compiled epidemiological data; N.M.-T. performed experiments; P.D., N.M.-T., M.L.G. and D.C. H. analyzed data; P.D., N.M.-T., O.S., D.C.H. and K.V.B. performed and/or supervised statistical studies; G.C. performed experiments; S.L., M.S. and E.K.R. reviewed and interpreted data.
Corresponding authors
Ethics declarations
Competing interests
The authors declares no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Methods, Supplementary Tables 1–5 and Supplementary Figures 1–23
Supplementary Dataset
Somatic mutation calls; list of somatic mutations detected in this study. Includes data for all mutated participants and time points
Rights and permissions
About this article
Cite this article
Desai, P., Mencia-Trinchant, N., Savenkov, O. et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat Med 24, 1015–1023 (2018). https://doi.org/10.1038/s41591-018-0081-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-018-0081-z
This article is cited by
-
Clonal hematopoiesis and its impact on the aging osteo-hematopoietic niche
Leukemia (2024)
-
Metabolic reprogramming regulated by TRAF6 contributes to the leukemia progression
Leukemia (2024)
-
Exploring the relationship between immune heterogeneity characteristic genes of rheumatoid arthritis and acute myeloid leukemia
Discover Oncology (2024)
-
Crosstalk between DNA methylation and hypoxia in acute myeloid leukaemia
Clinical Epigenetics (2023)
-
Recent advances in targeted therapies in acute myeloid leukemia
Journal of Hematology & Oncology (2023)