Abstract
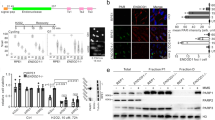
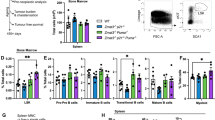
It has long been assumed that p53 suppresses tumor development through induction of apoptosis, possibly with contributions by cell cycle arrest and cell senescence1,2. However, combined deficiency in these three processes does not result in spontaneous tumor formation as observed upon loss of p53, suggesting the existence of additional mechanisms that are critical mediators of p53-dependent tumor suppression function3,4,5. To define such mechanisms, we performed in vivo shRNA screens targeting p53-regulated genes in sensitized genetic backgrounds. We found that knockdown of Zmat3, Ctsf and Cav1, promoted lymphoma/leukemia development only when PUMA and p21, the critical effectors of p53-driven apoptosis, cell cycle arrest and senescence, were also absent. Notably, loss of the DNA repair gene Mlh1 caused lymphoma in a wild-type background, and its enforced expression was able to delay tumor development driven by loss of p53. Further examination of direct p53 target genes implicated in DNA repair showed that knockdown of Mlh1, Msh2, Rnf144b, Cav1 and Ddit4 accelerated MYC-driven lymphoma development to a similar extent as knockdown of p53. Collectively, these findings demonstrate that extensive functional overlap of several p53-regulated processes safeguards against cancer and that coordination of DNA repair appears to be an important process by which p53 suppresses tumor development.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Cheok, C. F., Verma, C. S., Baselga, J. & Lane, D. P. Translating p53 into the clinic. Nat. Rev. Clin. Oncol. 8, 25–37 (2011).
Chan, T. A., Hwang, P. M., Hermeking, H., Kinzler, K. W. & Vogelstein, B. Cooperative effects of genes controlling the G2/M checkpoint. Genes Dev. 14, 1584–1588 (2000).
Brady, C. A. et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell 145, 571–583 (2011).
Li, T. et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 149, 1269–1283 (2012).
Valente, L. J. et al. p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma, and Noxa. Cell Rep. 3, 1339–1345 (2013).
Muller, P. A. & Vousden, K. H. p53 mutations in cancer. Nat. Cell Biol. 15, 2–8 (2013).
Vogelstein, B., Lane, D. & Levine, A. J. Surfing the p53 network. Nature 408, 307–310 (2000).
Lowe, S. W. et al. p53 status and the efficacy of cancer therapy in vivo. Science 266, 807–810 (1994).
Donehower, L. A. et al. Mice deficient for p53 are developmentally normal but are susceptible to spontaneous tumors. Nature 356, 215–221 (1992).
Jacks, T. et al. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4, 1–7 (1994).
Brady, C. A. & Attardi, L. D. p53 at a glance. J. Cell. Sci. 123, 2527–2532 (2010).
Kenzelmann Broz, D. et al. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 27, 1016–1031 (2013).
Jeffers, J. R. et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell 4, 321–328 (2003).
Villunger, A. et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 302, 1036–1038 (2003).
Deng, C., Zhang, P., Harper, J. W., Elledge, S. J. & Leder, P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 82, 675–684 (1995).
Michalak, E. M. et al. Puma and to a lesser extent Noxa are suppressors of Myc-induced lymphomagenesis. Cell Death Differ. 16, 684–696 (2009).
Valente, L. J., Grabow, S., Vandenberg, C. J., Strasser, A. & Janic, A. Combined loss of PUMA and p21 accelerates c-MYC-driven lymphoma development considerably less than loss of one allele of p53. Oncogene (2015).
Riley, T., Sontag, E., Chen, P. & Levine, A. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9, 402–412 (2008).
Lee, K., Tosti, E. & Edelmann, W. Mouse models of DNA mismatch repair in cancer research. DNA Repair (Amst) 38, 140–146 (2016).
Zhu, H. et al. Involvement of Caveolin-1 in repair of DNA damage through both homologous recombination and non-homologous end joining. PLoS ONE 5, e12055 (2010).
Razani, B. et al. Caveolin-1 null mice are viable but show evidence of hyperproliferative and vascular abnormalities. J. Biol. Chem. 276, 38121–38138 (2001).
Bersani, C., Xu, L. D., Vilborg, A., Lui, W. O. & Wiman, K. G. Wig-1 regulates cell cycle arrest and cell death through the p53 targets FAS and 14-3-3sigma. Oncogene 33, 4407–4417 (2014).
Harfe, B. D. & Jinks-Robertson, S. DNA mismatch repair and genetic instability. Annu. Rev. Genet. 34, 359–399 (2000).
Malkin, D. et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science 250, 1233–1238 (1990).
Schneider, K., Zelley, K., Nichols, K. E. & Garber, J. Li-Fraumeni Syndrome (GeneReviews, Seattle, WA, USA, 1999 [updated 2013]).
Sengupta, S. & Harris, C. C. p53: traffic cop at the crossroads of DNA repair and recombination. Nat. Rev. Mol. Cell Biol. 6, 44–55 (2005).
Lane, D. P. p53, guardian of the genome. Nature 358, 15–16 (1992).
Dudgeon, C. et al. The evolution of thymic lymphomas in p53 knockout mice. Genes Dev. 28, 2613–2620 (2014).
Sofer, A., Lei, K., Johannessen, C. M. & Ellisen, L. W. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol. Cell Biol. 25, 5834–5845 (2005).
Williams, A. B. & Schumacher, B. p53 in the DNA-damage-repair process. Cold Spring Harb. Perspect. Med. 6, a026070 (2016).
Dickins, R. A. et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat. Genet. 37, 1289–1295 (2005).
Aubrey, B. J. et al. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 10, 1422–1432 (2015).
Wu, X. et al. Dimerization of MLH1 and PMS2 limits nuclear localization of MutL. Mol. Cell. Biol. 23, 3320–3328 (2003).
Lois, C., Hong, E. J., Pease, S., Brown, E. J. & Baltimore, D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295, 868–872 (2002).
Herold, M. J., van den Brandt, J., Seibler, J. & Reichardt, H. M. Inducible and reversible gene silencing by stable integration of an shRNA-encoding lentivirus in transgenic rats. Proc. Natl Acad. Sci. USA 105, 18507–18512 (2008).
Strasser, A. et al. Enforced BCL2 expression in B-lymphoid cells prolongs antibody responses and elicits autoimmune disease. Proc. Natl Acad. Sci. USA 88, 8661–8665 (1991).
Dimri, G. P. et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. USA 92, 9363–9367 (1995).
Wang, H. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013).
Kueh, A. J. et al. An update on using CRISPR/Cas9 in the one-cell stage mouse embryo for generating complex mutant alleles. Cell Death Differ. 24, 1821–1822 (2017).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. https://arxiv.org/abs/1303.3997 (2013).
Josephidou, M., Lynch, A. G. & Tavaré, S. multiSNV: a probabilistic approach for improving detection of somatic point mutations from multiple related tumor samples. Nucleic Acids Res. 43, e61–e61 (2015).
Derrien, T. et al. Fast computation and applications of genome mappability. PLoS ONE 7, e30377 (2012).
Obenchain, V. et al. VariantAnnotation: a Bioconductor package for exploration and annotation of genetic variants. Bioinformatics 30, 2076–2078 (2014).
Willems, T. et al. Genome-wide profiling of heritable and de novo STR variations. Nat. Meth. 14, 590–592 (2017).
Scheinin, I. et al. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res. 24, 2022–2032 (2014).
Cameron, D. L. et al. GRIDSS: sensitive and specific genomic rearrangement detection using positional de Bruijn graph assembly. bioRxiv 110387 (2017).
Cameron, D. L. et al. GRIDSS: sensitive and specific genomic rearrangement detection using positional de Bruijn graph assembly. Genome Res. 27, 2050–2060 (2017).
Yin, T., Cook, D. & Lawrence, M. ggbio: an R package for extending the grammar of graphics for genomic data. Genome Biol. 13, R77 (2012).
Acknowledgements
We thank S. Lowe, A. Lujambio, A. Ventura and C. Concepcion for the p53 target gene shRNA library and for discussions; M. Ritchie, W. Shi, Y. Liao for data analysis, J.M. Adams, P. Bouillet, S. Cory, K. Rajewsky and A. Villunger for gifts of mice and for discussions. This work was supported by postdoctoral fellowships from the Leukemia & Lymphoma Society of America, Marie Curie Actions and Beatriu de Pinos, and Lady Tata Memorial Trust to A.J. and by grants from Cancer Australia and Cure Cancer Australia Foundation (grant no. 1067571), the National Health and Medical Research Council (NHMRC; program grant no. 1016701, NHMRC Senior Principal Research Fellow (SPRF) Fellowship 1020363 to A.S.), and the Leukemia & Lymphoma Society of America (Specialized Center of Research (SCOR) grant no. 7001-13), Australian Phenomics Network and a CCV Venture Grant. This work was made possible by operational infrastructure grants through the Australian Government Independent Medical Research Institutes Infrastructure Support Scheme (IRIISS) and the Victorian State Government Operational Infrastructure Support (OIS).
Author information
Authors and Affiliations
Contributions
The experiments were conceived and designed by A.J., M.J.H., L.O. and A.S. Experiments were performed mainly by A.J. with help from L.J.V., H.Y., L.T., L.M., S.W., C.J.V., S.M., A.J.K., M.S.B., L.M.L. and R.L.S. Whole genome sequencing analysis was performed by L.D.S., M.J.W. and A.T.P. The paper was written by A.J., M.J.H., L.O. and A.S. with help from the other authors.
Corresponding author
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–20 and Supplementary Tables 2–4
Supplementary Table 1
p53 gene targeted shRNA library
Rights and permissions
About this article
Cite this article
Janic, A., Valente, L.J., Wakefield, M.J. et al. DNA repair processes are critical mediators of p53-dependent tumor suppression. Nat Med 24, 947–953 (2018). https://doi.org/10.1038/s41591-018-0043-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-018-0043-5
This article is cited by
-
Combined absence of TRP53 target genes ZMAT3, PUMA and p21 cause a high incidence of cancer in mice
Cell Death & Differentiation (2024)
-
Alternative splicing and related RNA binding proteins in human health and disease
Signal Transduction and Targeted Therapy (2024)
-
Decoding p53 tumor suppression: a crosstalk between genomic stability and epigenetic control?
Cell Death & Differentiation (2024)
-
POLE2 knockdown suppresses lymphoma progression via downregulating Wnt/β-catenin signaling pathway
Molecular and Cellular Biochemistry (2024)
-
Transforming toxins into treatments: the revolutionary role of α-amanitin in cancer therapy
Archives of Toxicology (2024)