Abstract
Sepsis arises from diverse and incompletely understood dysregulated host response processes following infection that leads to life-threatening organ dysfunction. Here we showed that neutrophils and emergency granulopoiesis drove a maladaptive response during sepsis. We generated a whole-blood single-cell multiomic atlas (272,993 cells, n = 39 individuals) of the sepsis immune response that identified populations of immunosuppressive mature and immature neutrophils. In co-culture, CD66b+ sepsis neutrophils inhibited proliferation and activation of CD4+ T cells. Single-cell multiomic mapping of circulating hematopoietic stem and progenitor cells (HSPCs) (29,366 cells, n = 27) indicated altered granulopoiesis in patients with sepsis. These features were enriched in a patient subset with poor outcome and a specific sepsis response signature that displayed higher frequencies of IL1R2+ immature neutrophils, epigenetic and transcriptomic signatures of emergency granulopoiesis in HSPCs and STAT3-mediated gene regulation across different infectious etiologies and syndromes. Our findings offer potential therapeutic targets and opportunities for stratified medicine in severe infection.
Similar content being viewed by others
Main
The multiple dynamic host pathophysiological mechanisms that result in organ dysfunction following infection are incompletely understood and, while often aggregated into the clinical syndrome of sepsis, overlap with other critical illness syndromes1,2,3. There is an urgent need to better delineate such extreme responses to infection, given the COVID-19 pandemic and the wider, global burden of all-cause sepsis, which accounts for 11 million deaths per year and has a persistently high mortality of 20–30%4. Axes of immune dysregulation in sepsis have typically been examined by peripheral blood bulk transcriptomic studies, which lack the resolution to identify cell type-specific signatures5,6,7 or single-cell interrogation of peripheral blood mononuclear cells (PBMCs)8, which omits neutrophils (Neu).
Data from animal models demonstrate key protective and pathogenic roles for Neu in sepsis, in some instances involving specific subsets9,10. In humans, the clinical observation of a ‘left shift’ in complete blood count to increased immature Neu in severe infection is well recognized11. Neu abundance varies between septic and non-infectious inflammation12, while single-cell transcriptomics indicates that they are important in the pathogenesis of severe COVID-19 (ref. 13) and acute respiratory distress syndrome (ARDS)14. Expansion of subsets of immunosuppressive granulocytes associate with a higher risk of nosocomial infection15.
Sepsis is highly heterogeneous2 and immune suppression associated with apoptosis, epigenetic reprogramming and downregulation of activating cell surface molecules in multiple cell types2 is a predominant feature in many patients. Sepsis subphenotypes are reported2,5,6,7,16, but their relationship with mechanisms of immune dysfunction is not well defined. Sepsis response signatures (SRSs) from whole-blood transcriptomics identify dynamic response states, with assignments in the SRS1 group or a high likelihood of SRS1 group (SRSq), associated with immunosuppression, differential response to steroid therapy, more severe disease and higher early mortality5,17,18,19. Here we identified differences in Neu function and subset abundance during sepsis, which were a consequence of altered granulopoiesis. We demonstrated these immunosuppressive granulocytic and granulopoietic disturbances were the functional basis of the SRS1 subphenotype of sepsis.
Results
Immature and cycling neutrophils are increased in sepsis
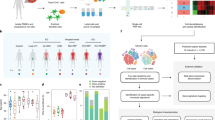
To generate an unbiased single-cell whole-blood (scWB) atlas of the sepsis response for all peripheral blood leukocyte populations, including Neu, we assayed freshly sampled whole blood from 26 patients with all-cause sepsis with a change in quick sequential organ failure assessment (SOFA) score of ≥2 points, indicating organ dysfunction and a physiological measure of acute illness severity score (national early warning score 2; NEWS2) of ≥7, indicating patients who required an urgent critical care response (Supplementary Table 1 and Methods); 9 sepsis convalescents (sampled 1–3 months after hospital discharge); 6 age- and sex-matched healthy controls (HCs); and 7 patients after cardiac surgery (CS) as a sterile inflammation control (Fig. 1a and Methods). We performed joint single-cell RNA and cell surface protein profiling of 272,993 cells (Fig. 1b, Extended Data Fig. 1 and Supplementary Table 2). After clustering and annotation of major immune cell types (Extended Data Fig. 1a–c and Methods), we observed recognized hallmarks of peripheral blood sepsis immunophenotypes, including neutrophilia, lymphopenia and reduced HLA-DR expression in CD14+CD16− classical monocytes (cMo)20 (Extended Data Fig. 1d).
a, Study design showing patients and analytical approaches applied. Discovery phase: scWB atlas for HCs (n = 6), sterile inflammation controls after CS (n = 7) and patients with sepsis (n = 26) by joint scRNA-seq and cell-surface protein profiling; scHSPC atlas for HCs (n = 7) and patients with sepsis (n = 15) (multiome scRNA-seq and scATAC-seq). Validation phase: mmV in HCs (n = 11) and patients with sepsis (n = 36), including CyTOF and bulk RNA-seq of whole blood and timsTOF proteomics in plasma; WB-CID datasets (n = 1,595) with deconvolution using scWB. b, Uniform Manifold Approximation and Projection (UMAP) for scWB (HCs, n = 6; CS, n = 7; sepsis, n = 26) scRNA-seq (272,993 cells) annotated for differential cell populations. DC, dendritic cell; cDCs, classical DCs; pDCs, plasmacytoid DCs. c, UMAP for scWB scRNA-seq differential abundance in samples from patients with sepsis (n = 26) compared to HCs (n = 11), with sampled neighborhoods colored by statistical significance (spatial FDR < 0.05). Nhood, neighborhood. d,e, Beeswarm plots of differential cell abundance in scWB with cluster labels of neighborhoods depicted and compared for patients with sepsis (n = 26) versus HCs (n = 11) (d) and patients with sepsis (n = 26) versus after CS (n = 7) (e).
We performed fine-resolution clustering and annotation (Extended Data Fig. 1e–i and Supplementary Table 3). RNA velocity and partition-based graph abstraction showed annotated Neu subpopulations followed the expected maturation sequence from immature MPO+ Neu to PADI4+ Neu to IL1R2+ Neu to S100A8/9+ Neu to mature CD10+CD16+ Neu (Extended Data Fig. 1j,k). We identified that degranulating CEACAM8+ Neu, S100A8/9hi Neu, IL1R2+ Neu, PADI4+ Neu, MPO+ Neu and cycling MK167+CYP1B1+ Neu were all proportionally increased in sepsis compared to HCs, whereas all mononuclear cell subsets, except CD19+CD38+CD71+ plasmablasts, were reduced (Fig. 1c,d). The increase in degranulating CEACAM8+ Neu and S100A8/9hi Neu and the reduction in all mononuclear cell subsets, was also seen in CS compared to HCs (Extended Data Fig. 2a,b), suggesting they represented nonspecific features of inflammation. By contrast, higher abundance of the immature IL1R2+ Neu, PADI4+ Neu and MPO+ Neu subsets and cycling MK167+CYP1B1+ Neu were specific to sepsis (Fig. 1c–e and Extended Data Fig. 2c,d). These findings demonstrated differential abundance of specific immature and cycling Neu subsets in sepsis.
Neutrophils in sepsis and recovery are immunosuppressive
To functionally test the immunosuppressive properties of sepsis Neu, we co-cultured bulk CD66b+ Neu (isolated from fresh whole blood of patients with sepsis by immunomagnetic selection) with allogeneic CD4+ T cells isolated from healthy donor leukocyte cones at a 4:1 Neu:T cell ratio in medium supplemented with interleukin (IL)-2 and CD3/CD28 Dynabeads. After 72–96 h of culture, the fraction of proliferating CD4+ T cells (calculated using non-bead-stimulated T cells cultured without Neu as the baseline proliferative fraction of cells in each sample) and the percentage of either PD-1+ or CD69+ CD4+ T cells (calculated relative to the CD3/28 bead-stimulated, no T cells control culture, to account for donor variation) was lower when co-cultured with CD66b+ Neu from patients with sepsis compared to HCs (Fig. 2a), indicating sepsis CD66b+ Neu inhibited CD4+ T cell proliferation and activation. There was no difference in the percentage of live, non-apoptotic CD4+ T cells on far-red DNA staining after 72–96 h of co-culture with either HCs or sepsis CD66b+ Neu (Extended Data Fig. 2e). For samples on which both single-cell sequencing and functional co-culture assays were performed, we correlated MPO+ Neu, PADI4+ Neu, IL1R2+ Neu, S100A8/9+ Neu, degranulating CEACAM8+ Neu and mature CD10+CD16+ Neu frequency with the fraction of proliferative CD4+ T cells, PD-1 expression and CD69 expression. None showed statistically significant correlation (Extended Data Fig. 2f), suggesting that all Neu subsets might be involved in the observed effect of CD66b+ Neu on CD4+ T cell proliferation and PD-1 and CD69 expression.
a, Percentage of eFluor450 cell proliferation dye+ CD4+ T cells (top) and activated PD-1+ (middle) and CD69+ (bottom) CD4+ T cells in co-cultures of bulk CD66b+ Neu isolated by immunomagnetic selection from whole blood of patients with sepsis (n = 22) or HCs (n = 10) and allogeneic CD4+ T cells from healthy donor leukocyte cones cultured at a 4:1 Neu:CD4+ T cell ratio in medium supplemented with IL-2 and CD3 + CD28 Dynabeads for 72–96 h relative to healthy CD4+ T cells cultured with beads and without Neu. Non-bead-stimulated T cells cultured without Neu were used as the baseline proliferative fraction of cells in each sample as a readout of proliferation. CD3 + CD28 bead-stimulated, no co-culture T cells were used to account for donor variation for PD-1+ and CD69+CD4+ T cells. Box plots denote minimum and maximum with whiskers and bottom quartile, median and upper quartile with the box. b, Heat maps showing DGE for mature CD10+CD16+ Neu, S100A8/9hi Neu, degranulating CEACAM8+ Neu and IL1R2+ Neu comparing patients with sepsis (n = 22) versus HCs (n = 10). c, Correlation (Pearson r, FDR < 0.05, 95% confidence interval) of days after acute sepsis and the proportion of mature CD10+CD16+ Neu in convalescent samples (n = 9) collected at 1–6 months after hospital discharge. d, Consensus DGE analysis stacked bar plots of all Neu in convalescent samples compared to all Neu in HC samples. Downreg, downregulated; upgreg, upregulated. e, Frequency of PD-1+ (left) and CD69+ (right) CD4+ T cells at 72–96 h of co-culture with bulk CD66b+ Neu isolated by immunomagnetic positive selection from whole blood from convalescent patients (n = 9) and HCs (n = 10). Box plots denote minimum and maximum with whiskers and bottom quartile, median and upper quartile with the box. f, Assessment of phagocytosis by ingestion of pHrodo Green Escherichia coli bioparticles stained with 7-AAD, CD66b-AF700 and Siglec-8-APC in bulk CD66b+ Neu isolated from convalescent patients (n = 9) and HCs (n = 10) as in e. Box plots denote minimum and maximum with whiskers and bottom quartile, median and upper quartile with the box. Conv, convalescence. Functional assays were tested with two-sided Wilcoxon rank-sum tests. *P < 0.05, **P < 0.01, ***P < 0.001.
Depletion of arginine by increased arginase-1 activity and upregulation of T cell immune checkpoint PD-1–PD-L1/PD-L2 pathways can modulate the immunosuppressive properties of granulocytic myeloid-derived suppressor cells (G-MDSCs)12,15. Inhibition with arginine and an Arg1 inhibitor, or antibodies to PD-L1/PD-L2 applied to the CD66b+ Neu-CD4+ T co-cultures, did not reverse the effects on proliferation, PD-1 and CD69 expression compared to co-cultures without inhibitors (Extended Data Fig. 2g). Addition of a prostaglandin EP2 receptor antagonist (TG6-10-1) increased CD69 expression compared to no inhibitor (Extended Data Fig. 2g), whereas the cyclo-oxygenase inhibitor indomethacin or a selective prostaglandin EP4 receptor competitive antagonist (GW-627368) did not (Extended Data Fig. 2g), suggesting effects at the level of pre-formed prostaglandin E2.
To understand how sepsis CD66b+ Neu mediated CD4+ T cell suppression, we analyzed differential gene expression (DGE) of mature CD10+CD16+ Neu, S100A8/9hi Neu, degranulating CEACAM8+ Neu and IL1R2+ Neu in the scWB dataset (Fig. 2b). We tested for and observed statistically significant enrichment of prostaglandin regulation or synthesis pathways in all tested Neu subsets from patients with sepsis compared to HCs (Extended Data Fig. 2h). We investigated the suppressive capacity by analyzing DGE indicative of experimentally validated G-MDSCs21,22,23 and found increased expression of this gene set (including SLC2A6, MMP8 and DUSP6) in both CD10+CD16+ Neu and IL1R2+ Neu and PADI4+ Neu and MPO+ Neu from patients with sepsis compared to HCs (Extended Data Fig. 2i).
To test whether the increased abundance of immature Neu populations in sepsis and their immunosuppressive features were linked to increased bone-marrow release of immature Neu, we analyzed samples at 1–6 months after hospital discharge (n = 9) (scWB cohort). Although no individual Neu subset was differentially abundant between convalescent and HC samples (Extended Data Fig. 3a), we observed a strong correlation in convalescent samples between time from acute sampling and increasing proportion of CD10+CD16+ Neu (Fig. 2c), suggesting that recovery involved gradual resumption of mature Neu production over 6 months. Consensus DGE of all Neu showed genes, including IL1B and type I interferon pathway (IFIT1, IFIT2, IFIT3, IFI6 and MX1) were downregulated in convalescence compared to HCs (Fig. 2d). Using the same cell isolation and co-culture system as for acute sepsis, we found convalescent CD66b+ Neu sampled at 1–6 months did not suppress CD4+ T cell proliferation (Extended Data Fig. 3b), but expression of CD69 and PD-1 on CD4+ T cells were reduced and phagocytosis was increased in sepsis convalescence samples compared to HCs (Fig. 2e,f). These observations demonstrated immunosuppressive features of sepsis Neu on CD4+ T cells, acutely and in convalescence, which involved multiple Neu subsets with a potential role for prostaglandins.
Sepsis alters granulopoietic profile of circulating HSPCs
To further investigate granulopoiesis, we performed single-cell RNA-seq (scRNA-seq) and single-cell assay for transposase-accessible chromatin with sequencing (scATAC-seq) on sorted live, singlet, CD34+CD45+ hematopoietic stem and progenitor cells (HSPCs) from PBMCs isolated from 15 patients with sepsis and 7 HCs. We assigned HSPC identity by mapping scRNA-seq data to healthy donor reference bone-marrow mononuclear cell scRNA-seq datasets24,25, performed clustering after multidimensional reduction by combining scRNA-seq and scATAC-seq, with assignment using majority RNA mapping (Extended Data Fig. 3c–f). Multimodal clustering identified five clusters of progenitor cells (Extended Data Fig. 3g). In patients with sepsis compared to HCs, cluster P4 (representing lymphocyte progenitors CDNTThi, FLT3hi, CD79hi and HOPXhi) was reduced (false discovery rate (FDR) = 0.002) (Extended Data Fig. 3h,i), whereas cluster P3 (defined by erythroid lineage progenitor genes HBBhi, HBDhi, KLF1hi and AC1hi) was increased (FDR = 0.0002) (Extended Data Fig. 3h,i), suggesting lymphoid depletion and an erythro-myeloid bias in sepsis.
Multimodal clustering on 29,336 hematopoietic stem cells (HSCs) retained after excluding progenitor cells identified seven clusters (Fig. 3a). Comparing patients with sepsis to HCs, clusters C3, C4, C5 and C7 were enriched in sepsis, whereas C6 was enriched in HCs (Fig. 3b and Extended Data Fig. 3j). To understand whether any expanded clusters represented HSCs with a granulopoietic bias, we leveraged Human Cell Atlas (HCA) annotated HSCs and progenitor scRNA-seq bone-marrow data26 to define genes upregulated in granulopoietic cells, including MPO, RNASE2, ELANE and FKBP2 (Methods and Extended Data Fig. 3k). Clusters C5 and C7 had the highest expression of this gene set (Fig. 3c). Granulopoiesis is driven by transcriptional circuits mediated by CEBP transcription factors, where CEBPA is important during steady-state granulopoiesis (SSG) and CEBPB is a key regulator of emergency granulopoiesis (EG)27,28. CEBPA/CEBPB binding motifs were enriched in accessible chromatin sites in both C5 and C7 (Fig. 3d), consistent with upregulated SSG and EG. Comparison of gene expression profiles of C3, C4, C5 or C7 with C6 identified genes, including SETBP1 and FLT3, which are involved in expanded myelopoiesis29,30, as upregulated in C5 and C7, respectively (Fig. 3e,f). These data provide evidence for altered granulopoiesis in sepsis that involved specific HSC clusters.
a, UMAP of scRNA-seq- and scATAC-seq-defined HSC clusters (29,336 cells) after exclusion of progenitor cells, with data arising from sorted live, singlet, CD34+CD45+ HSPCs from the PBMCs of patients with acute sepsis (n = 15), convalescent sepsis patients (n = 5) and HCs (n = 7). b, Differential abundance of HSCs between patients with sepsis and HCs as in a, with corresponding beeswarm plot (right) and UMAP visualization of sampled neighborhoods colored by statistically significant enrichment (spatial FDR < 0.05) (left). c, Granulopoiesis gene set scores per HSC cluster with differential cluster scoring tested with Kruskal–Wallis and post hoc Dunn’s tests. Violin plot with median, 95% confidence interval and interquartile range. ****FDR < 0.0001 comparing C5 or C7 versus other clusters. NS, not significant. d, chromVAR transcription factor motif z score deviation for CEBPA (left) and CEBPB (right) in HSC from acute sepsis samples (n = 15) and HCs (n = 7). e,f, DGE between cluster C5 versus C6 (e) and C7 versus C6 (f) in sepsis samples (n = 15).
Altered granulopoiesis drives a sepsis subphenotype
We next investigated whether the altered Neu-granulopoietic profile observed here was related to a previously identified SRS1 subphenotype in patients with sepsis5,17. Based on the expression of a seven-gene set (DYRK2, CCNB1IP1, TDRD9, ZAP70, ARL14EP, MDC1 and ADGRE3), the SRS1 subphenotype was assigned to 16 out of 26 patients in the scWB cohort (Extended Data Fig. 4a,b), in agreement with consensus or unsupervised clustering of the pseudobulked scRNA-seq data (Extended Data Fig. 4c–e). IL1R2+ Neu and cycling MK167+CYP1B1+ Neu were increased in patient samples assigned as SRS1 compared to non-SRS1, whereas mononuclear cells, including CD14+CD16− cMo, CD56+ natural killer (NK) and LTB+IL7R+memory CD4+ T cells, were depleted (Fig. 4a,b). Large numbers of differentially expressed genes were detected in mature CD10+CD16+ Neu (EBI3, MYOSLID and SLC1A3), S100A8/9hi Neu (SLC1A3, KLF14 and SGPP2), degranulating CEACAM8+ Neu (SFXN1, HS3ST3B1 and PDE4D) and IL1R2+ Neu (NAIP, IDI1 and CLEC4D) in samples assigned as SRS1 compared to non-SRS1 (Extended Data Fig. 5a), with a corresponding separation by SRS status on principal-component analysis (PCA) for these populations (Fig. 4c). By contrast, there were minimal differences in mononuclear cell subsets between SRS1 and non-SRS1 (Extended Data Fig. 5b), consistent with SRS groupings being driven by Neu. Cell surface expression of IL-1R2, measured by flow cytometry, showed a moderate correlation with expression of IL1R2 in CD66b+ Neu isolated from patient samples assigned SRS1 (Extended Data Fig. 5c). In co-cultures, SRS1 CD66b+ Neu suppressed CD4+ T cell activation more than non-SRS1 CD66b+ Neu (Fig. 4d), but not CD4+ T cell proliferation (Extended Data Fig. 5d). Moreover, SRS1 CD66b+ Neu displayed reduced phagocytosis compared to non-SRS1 (Fig. 4e), which was not restored by the addition of granulocyte–macrophage colony-stimulating factor (GM-CSF) (Extended Data Fig. 5e). These data indicated that SRS1 represented an immunosuppressed state driven, at least in part, by Neu dysfunction.
a, Differential cell abundance from whole-blood scRNA-seq UMAP with sampled neighborhoods colored by statistical significance (spatial FDR < 0.05) in patient samples in the scWB cohort (n = 26 patients with sepsis) assigned as SRS1 (n = 16) or non-SRS1 (n = 10). b, Beeswarm plot of differential cell abundance in scWB with cluster labels of neighborhoods depicted and compared for patients assigned as SRS1 and non-SRS1 as in a. c, First two principal components from PCA of pseudobulked Neu states colored by SRS assignment. d, Percentage of PD-1+ (top) and CD69+ (bottom) CD4+ T cells after 72–96 h of co-culture with CD66b+ Neu isolated from samples assigned as SRS1 (n = 6) and non-SRS1 (n = 13). Box plots denote minimum and maximum with whiskers and bottom quartile, median and upper quartile with the box. e, Assessment of phagocytosis in CD66b+ Neu isolated from samples assigned SRS1 (n = 15) and non-SRS1 (n = 14) and incubated with pHrodo Green E. coli bioparticles stained with 7-AAD, CD66b-AF700 and Siglec-8-APC. Box plots denote minimum and maximum with whiskers and bottom quartile, median and upper quartile with the box. Functional assays tested with two-sided Wilcoxon rank-sum tests. *P < 0.05.
To investigate this further in a multimodal validation (mmV) cohort, we reanalyzed whole-blood bulk RNA-seq and mass cytometry (CyTOF) immunophenotyping data31 in 42 samples from 36 individuals with all-cause sepsis and 11 age- and sex-matched HCs (Supplementary Table 1). SRS assignment of the 36 patients recapitulated higher early mortality in SRS1 compared to non-SRS1 patients (Extended Data Fig. 6a,b). We identified eight Neu clusters in the CyTOF dataset (Extended Data Fig. 6c–f). More immature CD64+CD10loCD16loCD15lo Neu and CD71hiCD38+Ki-67+ pro-Neu or CD71loKi-67+ pre-Neu were detected in patient samples assigned SRS1 compared to non-SRS1 (Extended Data Fig. 6g,h). Pseudotime trajectory analysis, which arranged cells in a progression of sequential maturation stages, demonstrated over-representation of SRS1 samples earlier in the trajectory (Extended Data Fig. 6i–l). PCA of CyTOF data showed separation of Neu subsets, but not mononuclear cell compartments, between samples assigned as SRS1 compared to non-SRS1 (Extended Data Fig. 6m), indicating changes in Neu subsets drove the patient SRS grouping.
To further investigate drivers of the SRS subphenotype, we reduced mmV RNA-seq dimensionality to 33 gene modules using weighted gene coexpression network analysis (WGCNA) and identified 13 differentially expressed modules between SRS1 and non-SRS1 assigned samples (FDR < 0.01) (Extended Data Fig. 7a). The SRS1 upregulated modules were enriched for gene expression signatures of differentiating Neu32. Module 10 (bone-marrow Neu and stage 1 differentiating Neu gene sets, including ALOX5, CYBB and LCN2) (Extended Data Fig. 7b) correlated with immature CD64+CD10loCD16loCD15lo Neu frequency (Extended Data Fig. 7c) and a gene expression signature defining IL1R2+ Neu (in the scWB cohort) including IL1R2, PFKFB2 and RETN genes (Extended Data Fig. 7d and Methods). To further validate enrichment of IL1R2+ Neu in SRS1, we performed cell type and cell state deconvolution on an independent total leukocyte microarray dataset5,17,18 consisting of 542 patients with all-cause sepsis, using the scWB single-cell multiomics dataset as a reference. We observed that IL1R2+ Neu and cycling MK167+CYP1B1+ Neu were increased in patient samples assigned as SRS1 compared to non-SRS1 (Extended Data Fig. 7e).
Next, we performed multiomics factor analysis (MOFA) to integrate cell cluster (CyTOF), gene module (RNA-seq) and plasma proteomic data (Extended Data Fig. 7f and Methods). Of all factors, 1 and 2 were most divergent between patient samples assigned as SRS1 compared to non-SRS1 (Extended Data Fig. 7g–k). The SRS1 direction in both factors was driven by immature Neu and progenitor Neu cell abundance (including CD71hiCD38+Ki-67+ pro-Neu, CD71loKi-67+ pre-Neu and CD64+CD10loCD16loCD15lo immature Neu), together with SRS1 upregulated gene modules and accounted for most variance in the cell and gene module datasets, but not proteomics (Extended Data Fig. 7i). Factor 5 was most strongly related to differences in plasma protein abundance, but showed no difference across SRS (Extended Data Fig. 7g–i), consistent with gene modules and cell clusters, rather than plasma proteins, being associated with SRS. Overall, the results show that differences in immature Neu populations, assayed using different modalities and Neu-granulopoietic dysfunction, were enriched in the SRS1 patient subphenotype.
STAT3 and granulopoiesis regulators define SRS1
Next we investigated the specific pathways and mediators that contributed to observed differences in the SRS1-associated Neu subsets. Consensus non-negative matrix factorization of Neu subsets in the scWB cohort identified gene expression programs (GEPs) specific to Neu populations (Neu-GEP) that positively correlated with SRSq, namely CD10+CD16+ Neu_program_3 (enriched for IL-6 activation and the JAK–STAT3 signaling pathway (IL-6–JAK–STAT3), prostaglandin (PG) synthesis and regulation, hereafter ‘PG’) and CD10+CD16+ Neu_program_8 (tumor necrosis factor (TNF) signaling via nuclear factor (NF)-κB, hereafter ‘TNF’); S100A8/9hi Neu_program_8 (IL-6–JAK–STAT3, PG); degranulating_CEACAM8+ Neu_program_9 (PG); PADI4 +Neu_program_7 (hypoxia); and IL1R2+ Neu_program_5 (metabolic regulator MTORC1 signaling, hypoxia) and IL1R2+ Neu_program_8 (TNF) (Fig. 5a; Supplementary Tables 3 and 4). We predicted master regulators per GEP and found granulopoiesis transcription factors, including CEBPB and STAT3 (CD10+CD16+ Neu_program_3, S100A8/9hi Neu_program_8 and IL1R2+ Neu_program_8) and CEBPA (S100A8/9hi Neu_program_8) were highly enriched (Fig. 5b). STAT3 and SPI1 were highly enriched for the CD64+CD10loCD371lo Neu correlated module 10 (mmV cohort) (Fig. 5c).
a, Heat map of Neu subset GEPs and correlation with SRSq (Spearman’s Rho on the right of the heat map, FDR < 0.05) in whole-blood samples from patients with sepsis (n = 26, scWB cohort). b, Transcription factor prediction analysis (top 50 genes per program) for CD10+CD16+ Neu_program_3, S100A8/9hi Neu_program_8 and IL1R2+ Neu_program_8 that positively correlated with SRSq, as in a. c, Transcription factor prediction analysis for bulk RNA-seq module 10 in whole-blood samples from patients with sepsis (n = 36, mmV cohort) using as input the top 1% genes showing correlation with module 10 eigengene. d, Expression of IL-6, G-CSF, GM-CSF and M-CSF in plasma from SRS1 and non-SRS1 patients with sepsis (n = 36, mmV cohort) (two-sided Wilcoxon rank-sum test). e,f, Representative live-cell imaging of Neu NETosis (e) and relative fluorescence intensity (f) in CD66b+ Neu from whole blood of SRS1 (n = 10) and non-SRS1 (n = 10) patient samples 4 h after stimulation with PMA and incubation with DNA-bound Cytotox Green reagent. Green fluorescence denotes cells undergoing NETosis. A total of 20 runs were performed (two-sided Wilcoxon rank-sum test). Box plots denote minimum and maximum with whiskers and bottom quartile, median and upper quartile with the box. NES, normalized enrichment score. *FDR < 0.05; *** FDR < 0.001.
Given the role of STAT3 in EG and enrichment for STAT3 in SRSq-correlated Neu-GEP, we investigated whether cytokines known to induce granulopoiesis and signal through STAT3 differed in plasma abundance (mmV cohort). Granulocyte colony-stimulating factor (G-CSF) and IL-6 were elevated in patient samples assigned as SRS1 compared to non-SRS1, whereas macrophage colony-stimulating factor (M-CSF) and GM-CSF showed no statistically significant difference (Fig. 5d). Consistent with G-CSF priming of Neu in cancer increasing NETosis33, we found that CD66b+ Neu from patient samples assigned as SRS1 underwent more NETosis than non-SRS1 samples on live-cell imaging for DNA-bound Cytotox Green reagent when stimulated with phorbol 12-myrisate 13-acetate (PMA) (Fig. 5e,f). These data demonstrated elevated STAT3-driven GEP in different SRS1-correlated Neu subsets and increased levels of circulating cytokines that signal through STAT3 and control granulopoiesis in SRS1 compared to non-SRS1 patients.
STAT3 processes in HSC cluster drive EG in SRS1
To test whether the SRS1 patient subphenotype represented a state of maladaptive granulopoiesis that involved heightened STAT3-mediated EG, we investigated whether there was any difference in circulating HSCs between SRS1 and non-SRS-1 subphenotypes, specifically the cell clusters differentially associated with granulopoiesis (C5 and C7), in 15 patients with sepsis from the single-cell hematopoietic stem and progenitor cell (scHSPC) atlas cohort. Cluster C5, but not C7, was enriched in patient samples assigned SRS1 compared to non-SRS1 (Fig. 6a,b and Extended Data Fig. 8a). We characterized the differentially accessible regions (DARs) of chromatin that defined the clusters and found 184 and 718 DAR in C5 and C7, respectively compared to other clusters (FDR < 0.05, fold change (FC) > 1.5) (Extended Data Fig. 8b,c and Supplementary Table 5). The chromatin profiles for C5 and C7 were enriched for publicly available myeloid progenitor cell chromatin profiles (C5, HSC multipotential progenitor; C7, megakaryocyte progenitor) (Extended Data Fig. 8d,e), suggesting their myelopoietic bias. CEBP motifs were identified within the DARs in both C5 and C7 (Extended Data Fig. 8f,g), whereas STAT motifs were enriched in C5, but not C7 DARs (Extended Data Fig. 8f,g). To differentiate which transcription factors governed the identities of clusters C5 and C7, we overlapped DARs with public chromatin immunoprecipitation (ChIP)-seq datasets to match transcription factor occupancy profiles. Enrichment of binding profiles for both CEBPA and CEBPB was detected in C7 (Extended Data Fig. 8h), whereas only CEBPB was enriched in C5, indicating that C5 was more biased toward EG while C5, but not C7, exhibited overlap with STAT3 binding profiles (Fig. 6c,d).
a,b Differential HSC abundance between patients with sepsis assigned as SRS1 (n = 6) or non-SRS1 (n = 9) assessed using density-based distribution UMAP visualization (a) and the corresponding beeswarm plots with sampled neighborhoods colored by statistically significant enrichment (spatial FDR < 0.05) (b). c, chromVAR transcription factor motif z score deviation for STAT3 in HSC from patients with sepsis (n = 15). d, ChIP-seq overlap analysis for differentially open peaks in cluster C5 versus other clusters. The GIGGLE score denotes a composite significance and effect size. e, UMAP of HSCs with supervised pseudotime trajectory from cluster C6 to cluster C5 showing STAT3 and CEBPB gene expression with pseudotime. f, Correlation of genes (left) and transcription factor (TF) motifs (right) along the C6–C5 pseudotime trajectory (Pearson’s r > 0.5, FDR < 0.05). g, UMAP of in silico effects of knockout (top) and overexpression (bottom) of STAT3 in clusters C5, C6 and C7 of HSCs from patients with sepsis (n = 15). Arrows display predicted changes in cell fate after gene of interest is perturbed.
To identify transcription factors relevant to HSC clusters, we integrated RNA and chromatin accessibility from the scHSPC data. We constructed supervised pseudotemporal trajectories from cluster C6 (enriched in HCs) to C5 (enriched in SRS1) and C7 (enriched in sepsis, but not SRS1) (Fig. 6e and Extended Data Fig. 9a). STAT3 and CEBPB expression increased along the C6–C5 trajectory (Fig. 6e), but not the C6–C7 trajectory (Extended Data Fig. 9b). We then identified transcription factor genes and chromatin transcription factor motifs that both changed along a trajectory and were correlated across modalities. While the C6–C5 trajectory involved CEBPB and STAT3 (Fig. 6f), the C6–C7 trajectory highlighted CEBPA (Extended Data Fig. 9c,d), indicating the role of C7 and C5 in SSG and EG, respectively and that STAT3-driven EG was increased in the SRS1 subphenotype.
To validate the importance of CEBPB–STAT3 and CEBPA in governing the identities of C5 and C7, respectively, transcription factor-mediated gene regulatory networks were constructed with CellOracle from RNA-seq and ATAC-seq HSC data for all HSC clusters. In silico knockout of CEBPA led to C7 loss of identity and transition toward C6, whereas C5 cells were not affected (Extended Data Fig. 9e), while in silico CEBPB knockout disrupted C5 cell differentiation (Extended Data Fig. 9e). In silico overexpression of CEBPA or CEBPB reversed the directions of cell fate transitions in C5 and C7, respectively (Extended Data Fig. 9e), suggesting that C7 was a SSG cluster and C5 was an EG cluster. In silico knockout or overexpression of STAT3 had similar effects as CEBPB (Fig. 6g), suggesting that STAT3 was driving EG. An independent methodology, using scRNA splicing information and vector field analysis of HSCs C5, C6 and C7, showed the same effects on EG and SSG reversal following in silico knockout of CEBPB, CEBPA and STAT3 (Extended Data Fig. 9f–j). These observations established C7 as a CEBPA SSG HSC cluster and C5 as a CEBPB–STAT3 EG cluster, with SRS1 enrichment of C5 highlighting EG in SRS1.
Dysregulated granulopoiesis associates with SRS across infections
We next investigated whether cellular (for example, IL1R2+ Neu expansion) and molecular (for example, STAT3 GEP) properties of the SRS1 patient subphenotype were seen across other severe infectious disease contexts. We reanalyzed publicly available bulk transcriptomics whole-blood cross-infectious disease (brWB-CID) datasets for four contexts: infectious organism (SARS-CoV-2 (ref. 31), n = 77 and influenza34, n = 109); source of infection (community acquired pneumonia (CAP), n = 438 and fecal peritonitis (FP), n = 229)17; clinical syndrome (ARDS35, n = 77); and age group (pediatric sepsis36, n = 106). All samples were assigned to SRS1 or non-SRS1 based on the expression of the seven-gene set. Cell type and state deconvolution demonstrated expansion of IL1R2+ Neu (Fig. 7a) and, to a lesser degree, expansion of cycling MK167+CYP1B1+ Neu (Extended Data Fig. 10a) in patient samples assigned as SRS1 compared to non-SRS1 for all contexts. DGE testing showed enrichment of a combined set of all GEPs from scWB Neu subsets involving STAT3 (CD10+CD16+ Neu_program_3, S100A8/9hi Neu_program_8 and IL1R2+ Neu_program_8) (denoted ‘STAT3_combined_program’) in patient samples assigned as SRS1 compared to non-SRS1 across all contexts (Extended Data Fig. 10b).
a, Frequency of IL1R2+ Neu following whole-blood bulk transcriptomics (brWB-CID) deconvolution in patients with sepsis infected with SARS-CoV-2 (n = 77), influenza (n = 109), CAP (n = 438), FP (n = 229), ARDS (n = 77) and pediatric septic shock (n = 106). Box plots denote minimum and maximum with whiskers and bottom quartile, median and upper quartile with the box. Two-sided Wilcoxon rank-sum test comparing SRS1 and non-SRS1 groups. b,c UMAP (b) and frequency of immature IL1R2+ Neu in samples assigned as SRS1 or non-SRS1 in adult ARDS patients (n = 9) (c) after reference mapping of scRNA-seq data from whole blood to reference single-cell atlas derived from the scWB showing. Box plots denote minimum and maximum with whiskers and bottom quartile, median and upper quartile with the box. d, Heat maps of expression of STAT3 GEPs (CD10+CD16+ Neu_program_3, S100A8/9hi Neu_program_8 and IL1R2+ Neu_program_8) in the corresponding Neu subsets from patients with ARDS (n = 9) with violin plots on each side. Two-sided Wilcoxon rank-sum test comparing SRS1 and non-SRS1 groups. e, Violin plots of expression of a combined set of GEPs involving STAT3 (CD10+CD16+ Neu_program_3, S100A8/9hi Neu_program_8 and IL1R2+ Neu_program_8) from scWB Neu subsets in pseudobulked Neu from patients with ARDS (n = 9) assigned as SRS1 or non-SRS1. Two-sided Wilcoxon rank-sum test comparing SRS1 and non-SRS1 groups. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
We also analyzed published whole-blood transcriptomic datasets of patients with ARDS14 (n = 9) or COVID-19 (ref. 13) (n = 8) at a single-cell resolution (scWB-CID) using scWB as a reference for cell type and cell state annotation (Fig. 7b). IL1R2+ Neu were expanded in patients with SRS1 subphenotype compared to non-SRS1 (Fig. 7c and Extended Data Fig. 10c). The STAT3 GEP (CD10+CD16+ Neu_program_3, S100A8/9hi Neu_program_8 and IL1R2+ Neu_program_8) showed enrichment in patient samples assigned as SRS1 compared to non-SRS1 in ARDS (Fig. 7d) and COVID-19 (Extended Data Fig. 10d). Total Neu, as defined by the original authors13,14, showed higher expression of the STAT3_combined_program genes in patient samples assigned as SRS1 compared to non-SRS1 in ARDS (Fig. 7e) and COVID-19 (Extended Data Fig. 10e) providing cross-validation. These analyses indicated that the biological basis of the SRS patient subphenotype was independent of infectious organism, source of infection, clinical syndrome and age.
Discussion
Here, we showed that Neu-granulopoietic disturbances in sepsis involved expansion of specific populations of immature Neu, suppression of CD4+ T cells in co-culture and altered granulopoiesis, and demonstrated these features were enriched in a subset of patients (SRS1). These results defined SRS1 as a specific immunocompromised disease endotype.
Our fresh whole-blood single-cell multiomic atlas, with no cellular enrichments or depletions, ensured faithful recapitulation of the sepsis cellular landscape. Previous single-cell -omic profiling focused on PBMCs, identifying an immature, bone-marrow-derived monocyte state (MS1), expanded in and predictive of sepsis8. The immature Neu populations defined here, in particular IL1R2+ Neu, exhibit similar gene expression profiles to MS1 cells8. Direct comparison between MS1 and IL1R2+ Neu could potentially reveal similar myelopoietic processes leading to their generation. The extent to which mobilization of IL1R2+ Neu may occur elsewhere in systemic inflammation, such as reported in mice37 and whether IL1R2+ Neu and other Neu subsets may drive immune suppression in these contexts, remains unclear.
While altered myelopoiesis in sepsis has been described in mice38,39 and modeled in vitro40 here we presented evidence for amplified granulopoiesis in humans during sepsis and specifically, a dysregulated form of EG in the SRS1 endotype. Our data, together with the reported increased risk of infections in patients with clonal hematopoiesis, including sepsis41, triangulate on the bone marrow as foundational for the maladaptive response to infection, with the caveat that our samples derive from circulating HSPCs rather than bone-marrow tissue.
Going forward, it will be important to understand determinants of differential bone-marrow responses to infection and the myelopoietic legacy of severe infection, for example, through trained immunity42 or hematopoietic exhaustion43. Published reports suggest that previous exposures and inflammatory comorbidity may be important in influencing subsequent myelopoietic responses to infection44. Meanwhile, our observations of persistent granulocytic alterations in convalescence, both phenotypic and functional, add to the evidence that infectious and inflammatory stimuli have long-lasting myelopoietic and therefore innate immune ramifications42,45. CEBPB has a key role in induction of trained immunity in HSCs46 and STAT3 drives specific immunosuppressive properties in MDSCs21. We found that STAT3-CEBPB-driven EG was pathognomonic of SRS1, with STAT3 underpinning the granulopoietic–granulocytic axis, raising the hypothesis that SRS1 represents a state of maladaptive innate immune reprogramming and memory, with a potential opportunity for manipulating STAT3 activation to alleviate sepsis- and SRS1-associated immunosuppression. For example, G-CSF and IL-6 show increased expression in SRS1, canonically signal through STAT3 and are key contributors to EG47,48 with inhibition specifically in patients with SRS1 or high SRSq subphenotypes a possible immunotherapeutic strategy1. This is further supported by Mendelian randomization work, where lower IL-6R expression associated with reduced mortality in sepsis49 and the therapeutic benefit of targeting IL-6 in severe COVID-19 (ref. 50).
Limitations of our study include the extent that our single-cell-analysis patient cohorts are fully representative of the breadth of the sepsis syndrome. Further work is needed to understand the differential immunosuppressive properties and role of prostaglandins in Neu, altered granulopoiesis through study of patient bone marrow and experimental gene manipulation of transcription factors to verify in silico knockouts. Relevant animal model and experimental medicine studies to manipulate candidate therapeutic targets are needed to better understand cytokine inhibition strategies.
Collectively, our work identified a common innate immune and hematopoietic axis that contributes to the maladaptive immune response to infection during sepsis and specifically a poor outcome, immunocompromised, extreme response SRS1 patient endotype, advancing opportunities for personalized medicine.
Methods
Study ethics, patient cohorts and sample sets
scWB atlas
Volunteers self-reporting as healthy (HCs) and with no history of infection in the past 14 d were recruited into the Genetic diversity and gene expression in white blood cells study following informed consent and under ethical approval (South Central Oxford REC B, reference 06/Q1605/55). Samples from patients with acute sepsis were collected from patients ≥18 years of age who were admitted to Oxford University Hospitals NHS Foundation Trust, UK. Patients were recruited from the intensive care unit (ICU) if they had symptoms and signs of established sepsis (suspected infection with an acute change in total SOFA score ≥2 points)3 or from the emergency department and medical wards if they had a change in quick SOFA score by ≥2 points and a NEWS2 (ref. 51) score ≥7 or intensive care review requested. Exclusion criteria were as previously reported in the UK Genomic Advances in Sepsis (GAinS) study (NCT00121196)5: patients or consultees unwilling or unable to give consent; advanced directive to withhold or withdraw life-sustaining treatment; admission for palliative care only; pregnancy and 6 weeks post-partum; or severe acquired immunodeficiency including systemic high-dose steroid therapy (prednisolone 0.5 mg kg−1 d−1 for 14 d or equivalent), HIV infection, known regular therapy with immunosuppressive agents such as azathioprine or neutrophil counts <1,000 ml−1 due to any cause, including metastatic disease and hematological malignancies or chemotherapy, but excluding severe sepsis and solid organ/bone-marrow transplant recipient receiving immunosuppressive therapy. Convalescent sepsis samples were collected 1–6 months after hospital discharge from individuals with acute sepsis samples already taken. Post-CS samples were collected from patients older than 18 years of age who were admitted to Oxford University Hospitals NHS Foundation Trust, UK. Patients were eligible if they were (1) undergoing cardiac bypass surgery, (2) required postoperative ICU stay and (3) did not have an infection before surgery. Exclusion criteria were identical to those for patients with sepsis. Patients with sepsis and CS were recruited into the Sepsis Immunomics Study following informed consent and under ethical approval (South Central Oxford REC C, reference:19/SC/0296) between May and November 2021. Sepsis samples were collected on days 1, 3 or 5 of hospital or ICU admission, whereas CS samples were collected 1 d after surgery. Written informed consent was obtained from adults or from personal/nominated consultees for patients lacking capacity, with retrospective consent obtained from the patient once capacity was regained.
scHSPC atlas
This included samples from 15 patients with sepsis and 7 age and sex-matched HCs recruited under the same studies as in scWB and mmV (see below), with one and eight acute sepsis samples overlapping with scWB and mmV, respectively and six acute sepsis samples not analyzed in either cohort. Eight convalescent sepsis samples were included. Of the HCs, five were the same HCs as in mmV. HCs and patients with sepsis were recruited into the same studies as described under whole-blood single-cell atlas31.
Bulk RNA-seq and single-cell RNA-seq WB cross-infectious disease datasets (brWB-CID/scWB-CID)
Publicly available data of infectious disease cohorts recruited with different clinical approaches were reanalyzed. Bulk transcriptomic data were obtained for adult sepsis based on microarray5,18 and bulk RNA-seq17, COVID-19 (ref. 31) (https://zenodo.org/record/6120249#.YrLY_OzML0o), influenza34 (GSE111368), CAP and FP (EGAD00001008730), ARDS35 (GSE65682) and pediatric septic shock36 (GSE13904). Single-cell transcriptomic data were reanalyzed for ARDS52 and COVID-19 (ref. 13).
Fresh whole-blood sample processing for single-cell sequencing
Blood samples were drawn into EDTA tubes (BD Biosciences) and processed within 1 h of collection. Then, 1 ml whole blood was lysed with 9 ml 1× eBioscience red blood cell lysis buffer (Thermo Fisher) twice and 200,000 cells were transferred for antibody staining.
Whole-blood single-cell RNA and cell surface protein profiling
scRNA and cell surface protein sequencing was performed with the BD Rhapsody platform (633731/633733, whole transcriptome assay (633801)) using 30 AbSeq antibodies (1 μl per antibody) (Supplementary Table 2). Cells were stained following the manufacturer’s recommendations before single-cell capture targeting 6,000 cells per sample. Reverse transcription, complementary DNA amplification and library construction (633801) were performed following the manufacturer’s recommendations in six batches. Libraries were sequenced on a NovaSeq6000 (Illumina).
Whole-blood single-cell multiomic analysis
Analysis was performed with v.4.0.0 R and Python 3.8.6.
Preprocessing and quality control
Gene expression data were aligned using STARsolo53 (v.2.7.9a) (GRCh38) and spliced and unspliced counts were produced. Unfiltered files were used for cell calling (emptyDrops function, DropletUtils (v.1.10.3)54) with a unique molecular identifier (UMI) threshold of 100 and FDR of 0.5%. AbSeq reads were trimmed with Trimmomatic (v.0.39) to the 12 bp UMI+ 36 bp AbSeq nucleotide sequence and aligned to an artificial reference of the AbSeq nucleotide sequences using STARsolo.
Cells expressing <100 or >4,000 genes, >10% mitochondrial reads, >2% hemoglobin reads or a log10(UMI per gene) <0.6 were removed. Genes expressed in <10 cells or with a total count <3 were removed. Scrublet (v.0.2.3) and doublet detect (v.3.0) were both used on default settings to remove doublets with automatic thresholds.
Normalization, dimensionality reduction and clustering
Data were log normalized and 4,000 highly variable genes (HVGs) were identified using the Seurat vst algorithm (scanpy v.1.7.2).
Multimodal dimensionality reduction was performed with TotalVI (scvi-tools v.0.10.0)55 on default settings with all 30 proteins and HVGs with each individual sample set as a batch (as all samples were processed separately).
Unsupervised clustering was performed on the 20 TotalVI latent dimensions (Seurat FindNeighbors (k = 30) and FindClusters (smart local moving algorithm)). Clustering resolution was evaluated by cluster neighborhood purity, cluster average silhouette width and a 30-iteration bootstrap to determine cluster stability with respect to sampling noise (bluster v.1.0). Additionally, we inspected top markers per cluster to match with known biology and understand potential value in merging versus splitting clusters. Combining these elements, the default clustering resolution 0.8 was chosen, with one immature neutrophil cluster split into two (MPO+ immature neutrophils/progenitors and PADI4+ immature neutrophils) based on resolution 1.1.
Cell annotation and RNA velocity/trajectory analysis
Cell clusters were merged for protein marker-based annotation of major known immune cell types at a broad level and kept at the clustering resolution of choice for fine annotation. Lineage assignment was confirmed with SingleR (v.1.6.1) assignments. Fine annotation was conducted by inspecting biologically meaningful gene markers for T cell and neutrophil populations (Supplementary Table 3).
RNA velocity and partition-based graph abstraction analysis of the neutrophils (without degranulating or apoptosing neutrophils) was performed with scVelo (v.0.2.3, default settings of stochastic model)56. Moments were estimated using TotalVI reduced dimensions.
scRNA-seq pseudobulk unsupervised analysis (consensus clustering, PCA, hierarchical clustering) and SRS assignment/scoring
Gene expression was aggregated per individual for all cells and normalized (EdgeR trimmed mean of M-values; TMM). The top 10% most variable genes by mean absolute deviation were taken for unsupervised analysis.
Consensus clustering was performed with the ConsensusClusterPlus package (v.1.54.0) (ConsensusClusterPlus function, 1,000 repetitions, pItem = 0.95, pFeature = 0.1, inner/finallinkage = ward.D2 and distance = euclidean). Unsupervised hierarchical clustering was performed with Ward’s linkage and Euclidean distance.
The relevant genes (DYRK2, CCNB1IP1, TDRD9, ZAP70, ARL14EP, MDC1 and ADGRE3) were used for sepsis sample SRS assignment and SRSq score calculation by SepstratifieR (v.0.0.0.9)57.
Differential abundance analysis
Cell type/state DA across conditions was identified by sampling neighborhoods of cells from a k-nearest neighbors (k-NN) graph and looking for enrichment of either condition in each neighborhood as implemented in MiloR58. The 20 batch-corrected latent dimensions from TotalVI were used for MiloR (v.0.99.19) k-NN graph construction (k = 30) and neighborhood indexing (proportion = 0.1). DA testing was performed with generalized linear models, including age and sex as covariates (neighborhoods significant if spatial corrected FDR < 0.05).
DGE analysis
For pseudobulk DGE, gene expression was aggregated per individual and per cell type/state into pseudobulks. Genes were filtered per pseudobulk based on minimum expression of n counts in at least X samples, where X was the smallest comparator group and n was defined for each pseudobulk based on histograms of logged count distributions. Pseudobulks were normalized by the EdgeR TMM method (v.3.30.3)59. DGE was performed with generalized linear models as implemented in EdgeR with age, sex and sequencing batch included in the model.
Consensus DGE for acute sepsis versus HC IL1R2+ immature neutrophils (as cells were too sparse to run pseudobulk DGE) and convalescent versus HC total neutrophils was performed as described in ref. 60. Genes were filtered for those that showed reproducible change in the same direction in a minimum of six samples (smallest comparator group size, HC = 6 samples).
Consensus non-negative matrix factorization of neutrophil gene expression
The 1,000 HVGs for each neutrophil state were selected (Seurat SelectIntegrationFeatures) followed by cNMF (v.1.2) as previously described (100 factorizations)61 for 4–15 GEPs. The final GEP number per neutrophil state was chosen based on a tradeoff between stability versus error as recommended by the original authors. Mean GEP usage per sample for each neutrophil state was correlated with sample SRSq scores with FDR adjustment. The top 50 genes per GEP were taken for downstream analysis.
Transcription factor prediction analysis
GEPs positively correlated with SRSq were used for TF prediction analysis with the Cytoscape (v.3.9.1) plugin iRegulon (v.1.3)62 (default settings). The same analysis was performed for the top 1% genes correlating with module 10 eigengene from mmV cohort RNA-seq WGCNA (below).
IL1R2 + neutrophil defining gene set
Genes specific to IL1R2+ neutrophils were identified with Wilcoxon tests (Seurat FindMarkers) for sepsis IL1R2+ neutrophils, filtering to only retain genes with adjusted P value <0.01 and average log2FC > 1.
Gene set and pathway enrichments
Gene sets for G-MDSCs22,23 were scored in the scWB neutrophils (Seurat AddModuleScore). Pathway analysis was performed against MSigDB Hallmark pathways and prostaglandin-related pathways from the MsigDB C2 curated gene sets in ClusterProfiler (v.4.1.4)63.
Neutrophil functional assays
Isolation of neutrophils from whole blood
Neutrophils were isolated from 5–10 ml whole blood from EDTA Vacutainer tubes (BD Biosciences) using EasySep HLA Chimerism Whole Blood CD66b positive selection kit (StemCell) following the manufacturer’s instructions.
Phagocytosis assay
Neutrophils were incubated for 20 min in complete medium with or without (fluorescence minus one) pHrodo Green E. Coli bioparticles (Invitrogen) (1 neutrophil:10 bioparticles). Cells were washed and stained for surface markers for 30 min with 7-AAD, CD66b-AF700 (G10F5) and Siglec-8-APC (7C9) antibodies from BioLegend and acquired with a BD LSRFortessa X-20 analyzer. The phagocytosis median fluorescence intensity for single 7-AAD−CD66b+Siglec-8− cells was determined by subtracting the median fluorescence intensity of the fluorescence minus one control sample.
Neutrophil-allogeneic CD4+ T cell co-culture
Cryopreserved CD4+ T cells from healthy donor leukocyte cones were thawed and stained with 10 μM eBioscience Cell Proliferation Dye eFluor450 (Invitrogen) following the manufacturer’s recommendations. T cells were co-cultured with CD66b+ neutrophils in 96-well U-bottom plates (200,000 cells per well) at 37 °C 5% CO2 at a 4:1 neutrophil to T cell ratio in complete medium supplemented with 50 IU ml−1 recombinant human IL-2 (BioLegend) and anti-CD3/CD28 Dynabeads (Gibco) added at a 1:1 bead:T cell ratio. As controls, T cells were plated without neutrophils in every run, with and without anti-CD3/CD28 beads. To reverse T cell inhibition, we tested co-cultures with 1 mM l-arginine (Sigma-Aldrich), 1 μM arginase-1 inhibitor CB-1158 (Fisher Scientific), 10 ng ml−1 anti-CD274 (eBioscience clone MIH1), 10 ng ml−1 anti-CD273 (PD-L2) (eBioscience clone MIH18), 20 μg ml−1 PGE2 (Sigma-Aldrich), 10 μM indomethacin (Sigma-Aldrich), 15 μM EP2 inhibitor TG6-10-1 (MedChemExpress) or 15 μM EP4 inhibitor GW-627368 (MedChemExpress).
After 72–96 h of co-culture, cells were stained for surface markers followed by annexin V and 7-AAD to exclude dead/apoptosing cells. Samples were analyzed using the BD LSRFortessa X-20. All antibodies were purchased from BioLegend unless otherwise stated: CD3-APC (UCHT1), CD4-BUV395 (BD Biosciences, SK3), CD66b-AF700 (G10F5), PD-1-PE (NAT105), CD69-PECy7 (FN50), 7-AAD, annexin V− FITC. T cells were gated as singlet CD66b−CD4+CD3+annexin V−7-AAD− cells. Proliferation analysis by dye dilution was established using the non-bead-stimulated T cells cultured without neutrophils as the baseline proliferative fraction of cells in each sample. The proliferative fraction and percentage of cells expressing PD-1 and CD69 were calculated as a percentage relative to the anti-CD3/28 bead-stimulated, no co-culture T cells control to account for donor variation.
Flow cytometry data analysis
Flow cytometry data were analyzed using FlowJo (v.10), GraphPad Prism (v.9) and R (v.4.0.0). The flow cytometry gating strategy for neutrophil functional assays is described in Supplementary Note 1.
NETosis assay
Neutrophils were plated at 20,000 cells per well (100 μl per well) in Ham’s F-12K medium (Gibco) using 96-well flat-bottom plates coated with 0.01% poly-l-ornithine solution (Sigma). Then, 250 nM IncuCyte Cytotox Green Dye was added to measure NETosis. Cells were treated with 100 nM PMA as per the manufacturer’s recommendations then imaged with the IncuCyte Live-Cell Analysis System for up to 4 h.
PBMC isolation and cryopreservation
PBMCs were isolated from sepsis (acute and convalescent) and HC whole blood using density gradient centrifugation with Leucosep tubes (Greiner) and lymphoprep (StemCell) and cryopreserved in 10% dimethylsulfoxide (Cell Signaling Technology).
CD34+ hematopoietic stem and progenitor cell enrichment
PBMCs from 15 patients with acute sepsis, 7 age and sex-matched HCs and 8 convalescent sepsis samples were used for CD34+ HSPC isolation. Three convalescent sepsis samples were removed after preprocessing and demultiplexing (below) (samples were judged as too long since the acute episode (>6 months past hospital discharge) by retrospective clinical evaluation). The 30 samples were processed in six batches, with each batch containing at least one sample from each comparator group.
Cryopreserved PBMCs were thawed and resuscitated (first wash with 25 U ml−1 benzonase (88700, Thermo Fisher)). Magnetic activated cell sorting for CD34+ enrichment (130-100-453, Miltenyi Biotec) was performed twice, according to the manufacturer’s instructions. Cells were stained at 4 °C with CD34-PE (clone AC136, Miltenyi Biotec) and CD45-FITC (clone HI30, BioLegend) and DRAQ7 (live/dead) dye (424001, BioLegend) and FACS-sorted for live, singlet, CD34+CD45+ HSPCs.
HSPC single-cell multiomics
scRNA and scATAC-seq on CD34+ HSPCs was performed by isolating nuclei of flow-sorted HSPCs. An equal number of cells per sample was pooled for each batch. Cell lysis and nuclei extraction were conducted following the low input workflow within the 10x Genomics Demonstrated Protocol (CG000365 Rev B). Nuclei were transposed, captured and RNA and ATAC library preparation (1000285, 10x Genomics) was performed as per the 10x Genomics manufacturer’s protocol. RNA libraries were sequenced on a NovaSeq6000 (Illumina) and ATAC libraries were sequenced on a NextSeq500 (Illumina).
HSPC single-cell multiomic analysis
Analysis was performed with v.4.1.0 R using ArchR (v.1.0.2) unless otherwise specified64.
Preprocessing, demultiplexing and doublet removal
Raw FASTQ files of scRNA-seq and scATAC-seq were aligned to the GRCh38 reference genome using 10x CellRanger ARC (v.2). Genetic demultiplexing and doublet removal was performed for each batch as previously described31. HSPC multiomic data were input into ArchR (v.1.0.2) with mintss = 4 and minFrags = 1,000. Homotypic doublets were removed (removeDoublet).
Quality control
Cells expressing <100 or >6,000 genes, >25,000 UMIs or with a log10(UMI per gene) <0.8 were removed. Cells with TSS enrichment <7 and <1,000 unique fragments were also filtered out. Genes expressed in <10 cells or <3 in total count were removed. This left 46,782 cells (median TSS enrichment of 14.6, median fragment count of 13,821) and 26,660 genes.
Cell type/state annotations and non-HSC filtering
We assigned identity of HSPCs and contaminating cells by mapping our scRNA-seq data to two healthy donor reference bone-marrow mononuclear cell scRNA-seq datasets from ref. 24 (reference dataset 1, supervised PCA) and ref. 25 (reference dataset 2, PCA) with Seurat (v.4.0). Cells with non-HSPC labels from either mapping were filtered out, leaving 46,156 HSPCs.
Multimodal dimensionality reduction for HSPCs was performed (iterativeLSI) for scRNA (gene expression matrix) and scATAC (tile matrix). Both modalities were batch-corrected (Harmony) and combined (addCombinedDims) before clustering on the combined dimensions (resolution of 0.4, smart local moving algorithm). HSPC identity was finalized by assigning the majority RNA mapping identity from reference dataset 1 per cluster. Progenitor cells were thus filtered out and only 29,336 HSCs retained for downstream analysis.
Progenitor cell differential abundance analysis
Hematopoietic progenitor cells from the above filtering step (n = 16,820) were compared across sepsis and HC conditions by two-sided Wilcoxon rank-sum tests with FDR adjustment for multiple testing to look for differential abundance.
HSC peak calling and reclustering
Each individual sample was pseudobulked for peak calling with MACS2 and iterative peak overlapping removal65 within ArchR (minimum cells = number of cells in the sample with the fewest cells, minimum replicates = 5 (lowest sample group size that is convalescent sepsis samples)). We re-performed dimensionality reduction and batch correction on the gene expression and peak matrices before reclustering HSCs on combined dimensions (smart local moving algorithm, resolution = 0.2 to avoid overclustering and challenging interpretability given the purity of the cell type already). Cluster C2 was derived from only one single sample.
HSC differential abundance analysis
HSC cluster DA changes across conditions were conducted in MiloR (v.1.2.0). The 60 batch-corrected reduced dimensions (30 each of scRNA/scATAC) were used in MiloR k-NN graph construction (k = 20) and neighborhood indexing (proportion = 0.3). DA testing was performed with generalized linear models, including age and sex as covariates (neighborhoods were significant if spatially corrected FDR < 0.05).
Granulopoiesis gene set enrichment scoring for single cells
We reanalyzed HCA HSPC scRNA-seq data, utilizing annotations from Hay et al.26 to contrast all granulocytic progenitors with HSCs, multilineage progenitors, monocyte/DC progenitors, lymphoid-primed multipotent progenitors and megakaryocyte/erythroid progenitors to define a granulopoietic gene set. We scored our HSCs with this gene set (Seurat AddModuleScore) and compared differential gene set activity across HSC clusters with a Kruskal–Wallis test and post hoc Dunn’s test with FDR adjustment.
DGE and chromatin accessibility analysis
Genes defining the C5 and C7 HSC clusters (versus C6) were identified with Wilcoxon tests (getMarkerFeatures) with thresholds of FDR < 0.05 and FC > 1.5. Genomic regions defining the C5 and C7 HSC clusters were identified with Wilcoxon tests (getMarkerFeatures) with thresholds of FDR < 0.05 and FC > 1.5.
Bulk ATAC/TF motif enrichment analysis and ChIP-seq dataset overlapping
Differentially open peak regions for clusters C5 and C7 were taken for bulk ATAC/TF motif enrichment analyses (peakAnnoEnrichment). We also input the differentially open peak regions into the cistromeDB toolkit (http://dbtoolkit.cistrome.org/)66 (top 1,000 peaks according to peak enrichment used).
Integrative trajectory analysis
Pseudotemporal trajectories was constructed (addTrajectory) on the combined batch-corrected LSI dimensions with clusters C6–C5 (enriched in HC/SRS1, respectively) and C6–C7 (enriched in HC/sepsis, respectively) as the backbone. Trajectories for both gene expression and TF motifs were extracted (getTrajectory) and correlated (correlateTrajectories, maxDist 500 kb).
Gene regulatory network construction and in silico knockout/overexpression analyses
We built gene regulatory networks (GRNs) with CellOracle (v.10.10)67 following tutorials from https://morris-lab.github.io/CellOracle.documentation/. In brief, HSPC scATAC data were used to build the base GRN via Cicero68, following which, scRNA data were used to prune the GRN though a Bagging Ridge model. Instead of recomputing principal components for the scRNA data for GRN construction, we instead used the batch-corrected joint ATAC and RNA LSI dimensions. Effects on gene expression of HSC clusters C5/6/7 when perturbing specific TFs (CEBPB, CEBPA and STAT3) were simulated by setting expression of the TF either to 0 or double the value of its maximum expression, with CellOracle then predicting based on the new simulated gene expression changes the trajectory of cellular transition.
RNA velocity and vector field analyses
We recapitulated expression dynamics vector fields with dynamo (v.1.1.0 (ref. 69)) following tutorials from https://dynamo-release.readthedocs.io/en/latest/. In brief, HSPC scRNA raw sequencing data that are FASTQ files were reprocessed by STARsolo (v.2.7.9a) (GRCh38) and spliced and unspliced counts produced for the already identified HSPCs. RNA velocity, acceleration and curvature were then calculated for clusters C5/6/7. Effects on RNA velocity on HSC clusters C5/6/7 when perturbing specific TFs (CEBPB, CEBPA and STAT3) were simulated by setting expression of the TF to 0. The effect of a change in expression of these TFs on the regulation (activation versus inhibition) of each other was also analyzed by calculating the RNA Jacobian for the HSCs.
SRS assignment for neutrophil functional assay samples and scHSPC samples not part of scWB or mmV cohorts
Whole blood was sampled into Tempus tubes (Thermo Fisher) and stored at −80 °C until RNA extraction with Norgen Preserved Blood RNA Purification kit I (43400) according to the manufacturer’s instructions. RNA was used for quantitative PCR with reverse transcription for the seven SRS genes and Cq values input into SepstratifieR for SRS assignment as previously described57.
Bulk RNA-seq analysis
Analysis was performed with v.4.0.0 R. The EdgeR TMM normalized count matrix was obtained from the COMBAT consortium31 and SRS assignment was conducted as for scWB samples.
WGCNA
WGCNA (v.1.70-3) was conducted as per the vignettes from https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/Tutorials/ with the log normalized count matrix. A signed-hybrid network (soft threshold of 4) was constructed with biweight mid-correlation. Minimum module size was 30. Modules with eigengene correlation <0.2 were merged, leaving 33 modules.
Pathway analysis
Pathway analysis was performed against neutrophil gene sets from the HCA26 and signatures of differentiating neutrophils from mouse single-cell transcriptomics32 (ClusterProfiler enricher). Gene set enrichment analysis for scWB IL1R2+ neutrophil defining genes was performed via the gene set enrichment analysis function against all genes in module 10 ranked by their correlation with the module eigengene.
CyTOF analysis
Analysis was performed with v.4.0.0 R.
Preprocessing
Normalized, debarcoded, bead and doublet-cleaned FCS files were obtained from the COMBAT consortium31. To avoid biases due to highly varying cell numbers per sample, a maximum of 50,000 cells per sample was taken. Harmony (v.1.0)70 was used for batch correction71.
Clustering, trajectory inference and differential abundance analyses
Clustering and consensus clustering were performed with the CATALYST package (v.1.14.0)72,73 with all cells and 41 markers. The 50 metaclusters were merged following manual annotation. As the 41-marker panel was myeloid focused, we annotated five monocyte clusters, before subsetting the neutrophils and reclustering with 17 selected markers. Thirty neutrophil metaclusters were manually annotated and merged based on median marker expression74 into eight subsets, leaving a final total of 22 clusters at the finest level of annotation (14 non-neutrophil and eight neutrophil).
For trajectory analysis, neutrophils were downsampled to 15,000 cells per comparator group for CATALYST diffusion map dimensionality reduction followed by principal curve fitting (Slingshot v.1.8.0, default settings)75.
PCA of cluster proportions was performed after quantile normalization. Cluster DA analysis with FDR adjustment was performed with generalized linear mixed models (glmmPQL function, MASS package, v.7.3-51.6) to account for repeated sampling in six patients, with batch included as an additional fixed effect and patient a random effect. An extra random effect term per sample was included to model overdispersion in proportions seen in high dimensional cytometry data73.
Multimodal analysis by MOFA+
We integrated mmV bulk RNA-seq and CyTOF data with MOFA+ (v.1.0.1)76, including 105 plasma protein measurements (timsTOF mass spectrometry) from the same patient samples31 as a computational negative control, reasoning that the latent spaces linking cell clusters to gene expression profiles should not show an overly strong contribution from the plasma proteome, which reflects contributions from various tissues, whereas the RNA was derived solely from leukocytes. For each sample, the eigengenes of the 33 identified modules from WGCNA, quantile normalized proportion of the 22 cell clusters from CyTOF and log2-transformed and median centered intensity values for 105 plasma proteins were utilized. Views were left unscaled and all other model options were left with default values.
Cytokine analysis
Concentrations of plasma analytes from the Luminex assay were obtained from the COMBAT consortium31.
brWB-CID bulk transcriptomic analysis
A signature matrix for CIBERSORTx77 was constructed using our scWB single-cell dataset, with each finely annotated population except apoptosing cells downsampled to 100 cells per population. The signature matrix was then created via the Create Signature Matrix analysis module with min.expression = 0.25, replicates = 100 and sampling = 0.5.
SRS was assigned for the bulk transcriptomic data of sample set 3 as described for cohorts 1 and 2. Cell fractions of the 1,578 whole-blood bulk transcriptomic samples of sample set 3 were then estimated with the Impute Cell Fractions analysis module using the single-cell reference matrix, with batch correction S-mode enabled quantile normalization disabled for RNA-seq datasets. DGE was performed across SRS for each cohort and SRS1 upregulated genes were tested for enrichment of the union of genes of all three STAT3 GEPs from scWB neutrophil cNMF.
scWB-CID scRNA-seq analysis
ARDS52 and COVID-19 (ref. 13) whole-blood scRNA-seq data were mapped to the scWB data (Seurat FindTransferAnchors and MapQuery) and cell type/state proportions examined. We then scored the neutrophils for scWB neutrophil cNMF STAT3 GEPs (Seurat AddModuleScore).
Statistics and reproducibility
In addition to the above details of statistical methods, we ensured analytical rigor with the following procedures.
For scWB, we estimated from existing data5,18 that the smallest SRS group would be one-third of the recruited patients with sepsis; we opted to recruit 26 patients for a minimum of 8 patients in the smallest group. No sample size calculations were performed. Cells from all study participants were used to determine cell states. For scHSPC, three convalescent sepsis samples were excluded (above), after which all 27 samples were used for all analysis. Sample size was determined to be adequate based on the degree and consistency of differences between groups.
For scWB and scHSPC, sequencing batches were prepared such that each batch contained samples from all comparator groups (acute sepsis, HCs, after CS and convalescent sepsis). SRS groups were not known at the time of patient recruitment and data generation.
Cells from all samples in scWB, mmV CyTOF and scHSPC were analyzed blind to which patient they originated from to define the varying cell states.
Data distributions were assumed to be normal for (generalized) linear models but this was not formally tested. Box plots denote minimum and maximum with whiskers and bottom quartile, median and upper quartile with the box.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Raw data for whole-blood single-cell sequencing and HSC single-cell sequencing datasets are deposited on the European Genome–phenome Archive (EGAS00001006283) and derived data are at Zenodo (https://doi.org/10.5281/zenodo.7723202). For sequence-level raw datasets deposited at the European Genome–phenome Archive, access is managed by a Data Access Committee.
Code availability
Code used for every algorithm followed in data processing and analysis is fully referenced within the specific Methods sections.
References
Maslove, D. M. et al. Redefining critical illness. Nat. Med. 28, 1141–1148 (2022).
van der Poll, T., Shankar-Hari, M. & Wiersinga, W. J. The immunology of sepsis. Immunity 54, 2450–2464 (2021).
Singer, M. et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 315, 801–810 (2016).
Rudd, K. E. et al. Global, regional, and national sepsis incidence and mortality, 1990-2017: analysis for the Global Burden of Disease Study. Lancet 395, 200–211 (2020).
Davenport, E. E. et al. Genomic landscape of the individual host response and outcomes in sepsis: a prospective cohort study. Lancet Resp. Med. 4, 259–271 (2016).
Scicluna, B. P. et al. Classification of patients with sepsis according to blood genomic endotype: a prospective cohort study. Lancet Resp. Med. 5, 816–826 (2017).
Sweeney, T. E. et al. Unsupervised analysis of transcriptomics in bacterial sepsis across multiple datasets reveals three robust clusters. Crit. Care Med. 46, 915–925 (2018).
Reyes, M. et al. An immune-cell signature of bacterial sepsis. Nat. Med. 26, 333–340 (2020).
Qi, X. et al. Identification and characterization of neutrophil heterogeneity in sepsis. Crit. Care 25, 50 (2021).
Shen, X., Cao, K., Zhao, Y. & Du, J. Targeting neutrophils in sepsis: from mechanism to translation. Front. Pharm. 12, 644270 (2021).
Farkas, J. D. The complete blood count to diagnose septic shock. J. Thorac. Dis. 12, S16–S21 (2020).
Meghraoui-Kheddar, A. et al. Two new immature and dysfunctional neutrophil cell subsets define a predictive signature of sepsis useable in clinical practice. Am. J. Respir. Crit. Care Med. 205, 46–59 (2020).
Schulte-Schrepping, J. et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell 182, 1419–1440 (2020).
Sinha, S. et al. Dexamethasone modulates immature neutrophils and interferon programming in severe COVID-19. Nat. Med. 28, 201–211 (2022).
Uhel, F. et al. Early expansion of circulating granulocytic myeloid-derived suppressor cells predicts development of nosocomial infections in patients with sepsis. Am. J. Respir. Crit. Care Med. 196, 315–327 (2017).
Reddy, K. et al. Subphenotypes in critical care: translation into clinical practice. Lancet Resp. Med. 8, 631–643 (2020).
Cano-Gamez, E. et al. An immune dysfunction score for stratification of patients with acute infection based on whole-blood gene expression. Sci. Transl. Med. 14, eabq4433 (2022).
Burnham, K. L. et al. Shared and distinct aspects of the sepsis transcriptomic response to fecal peritonitis and pneumonia. Am. J. Respir. Crit. Care Med. 196, 328–339 (2017).
Antcliffe, D. B. et al. Transcriptomic signatures in sepsis and a differential response to steroids. From the VANISH randomized trial. Am. J. Respir. Crit. Care Med. 199, 980–986 (2019).
Cazalis, M. A. et al. Decreased HLA-DR antigen-associated invariant chain (CD74) mRNA expression predicts mortality after septic shock. Crit. Care 17, R287 (2013).
Veglia, F., Perego, M. & Gabrilovich, D. Myeloid-derived suppressor cells coming of age. Nat. Immunol. 19, 108–119 (2018).
Bayik, D. et al. Myeloid-derived suppressor cell subsets drive glioblastoma growth in a sex-specific manner. Cancer Discov. 10, 1210–1225 (2020).
Alshetaiwi, H. et al. Defining the emergence of myeloid-derived suppressor cells in breast cancer using single-cell transcriptomics. Sci. Immunol. 5, eaay6017 (2020).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Granja, J. M. et al. Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat. Biotechnol. 37, 1458–1465 (2019).
Hay, S. B., Ferchen, K., Chetal, K., Grimes, H. L. & Salomonis, N. The Human Cell Atlas bone marrow single-cell interactive web portal. Exp. Hematol. 68, 51–61 (2018).
Hirai, H. et al. C/EBPβ is required for ‘emergency’ granulopoiesis. Nat. Immunol. 7, 732–739 (2006).
Manz, M. G. & Boettcher, S. Emergency granulopoiesis. Nat. Rev. Immunol. 14, 302–314 (2014).
Böiers, C. et al. Expression and role of FLT3 in regulation of the earliest stage of normal granulocyte-monocyte progenitor development. Blood 115, 5061–5068 (2010).
Makishima, H. et al. Somatic SETBP1 mutations in myeloid malignancies. Nat. Genet. 45, 942–946 (2013).
COMBAT Consortium. A blood atlas of COVID-19 defines hallmarks of disease severity and specificity. Cell 185, 916–938 (2022).
Giladi, A. et al. Single-cell characterization of haematopoietic progenitors and their trajectories in homeostasis and perturbed haematopoiesis. Nat. Cell Biol. 20, 836–846 (2018).
Demers, M. et al. Cancers predispose neutrophils to release extracellular DNA traps that contribute to cancer-associated thrombosis. Proc. Natl Acad. Sci. USA 109, 13076–13081 (2012).
Dunning, J. et al. Progression of whole-blood transcriptional signatures from interferon-induced to neutrophil-associated patterns in severe influenza. Nat. Immunol. 19, 625–635 (2018).
Bos, L. D. J. et al. Understanding heterogeneity in biologic phenotypes of acute respiratory distress syndrome by leukocyte expression profiles. Am. J. Respir. Crit. Care Med. 200, 42–50 (2019).
Wong, H. R. et al. Identification of pediatric septic shock subclasses based on genome-wide expression profiling. BMC Med. 7, 34 (2009).
Martin, P. et al. Mouse neutrophils express the decoy type 2 interleukin-1 receptor (IL-1R2) constitutively and in acute inflammatory conditions. J. Leukoc. Biol. 94, 791–802 (2013).
Weber, G. F. et al. Interleukin-3 amplifies acute inflammation and is a potential therapeutic target in sepsis. Science 347, 1260–1265 (2015).
Kwok, I. et al. Combinatorial single-cell analyses of granulocyte-monocyte progenitor heterogeneity reveals an early uni-potent neutrophil progenitor. Immunity 53, 303–318 (2020).
Reyes, M. et al. Plasma from patients with bacterial sepsis or severe COVID-19 induces suppressive myeloid cell production from hematopoietic progenitors in vitro. Sci. Transl. Med. 13, eabe9599 (2021).
Zekavat, S. M. et al. Hematopoietic mosaic chromosomal alterations increase the risk for diverse types of infection. Nat. Med. 27, 1012–1024 (2021).
Mitroulis, I. et al. Modulation of myelopoiesis progenitors is an integral component of trained immunity. Cell 172, 147–161 (2018).
Pietras, E. M. et al. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat. Cell Biol. 18, 607–618 (2016).
Li, X. et al. Maladaptive innate immune training of myelopoiesis links inflammatory comorbidities. Cell 185, 1709–1727 (2022).
Naik, S. & Fuchs, E. Inflammatory memory and tissue adaptation in sickness and in health. Nature 607, 249–255 (2022).
de Laval, B. et al. C/EBPβ-dependent epigenetic memory induces trained immunity in hematopoietic stem cells. Cell Stem Cell 26, 793 (2020).
Zhang, H. et al. STAT3 controls myeloid progenitor growth during emergency granulopoiesis. Blood 116, 2462–2471 (2010).
Walker, F. et al. IL6/sIL6R complex contributes to emergency granulopoietic responses in G-CSF- and GM-CSF-deficient mice. Blood 111, 3978–3985 (2008).
Hamilton, F. et al. Therapeutic potential of IL6R blockade for the treatment of sepsis and sepsis-related death: findings from a Mendelian randomisation study. PLoS Med. 20, e1004174 (2023).
The REMAP-CAP Investigators. Interleukin-6 receptor antagonists in critically ill patients with COVID-19. N. Engl. J. Med. 384, 1491–1502 (2021).
Mellhammar, L. et al. NEWS2 is superior to qSOFA in detecting sepsis with organ dysfunction in the emergency department. J. Clin. Med. 8, 1128 (2019).
Sinha, S. et al. Dexamethasone modulates immature neutrophils and interferon programming in severe COVID-19. Nat. Med. 28, 201–211 (2022).
Kaminow, B., Yunusov, D. & Dobin, A. STARsolo: accurate, fast and versatile mapping/quantification of single-cell and single-nucleus RNA-seq data. Preprint at bioRxiv https://doi.org/10.1101/2021.05.05.442755 (2021).
Lun, A. T. L. et al. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 20, 63 (2019).
Gayoso, A. et al. Joint probabilistic modeling of single-cell multi-omic data with totalVI. Nat. Methods 18, 272–282 (2021).
Bergen, V., Lange, M., Peidli, S., Wolf, F. A. & Theis, F. J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38, 1408–1414 (2020).
Cano-Gamez, E. et al. An immune dysfunction score for stratification of patients with acute infection based on whole blood gene expression. Sci. Transl. Med. 14, eabq4433 (2022).
Dann, E., Henderson, N. C., Teichmann, S. A., Morgan, M. D. & Marioni, J. C. Differential abundance testing on single-cell data using k-nearest neighbor graphs. Nat. Biotechnol. 40, 245–253 (2022).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Wilk, A. J. et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 26, 1070–1076 (2020).
Kotliar, D. et al. Identifying gene expression programs of cell-type identity and cellular activity with single-cell RNA-Seq. eLife 8, e43803 (2019).
Janky, R. et al. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput. Biol. 10, e1003731 (2014).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Granja, J. M. et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet. 53, 403–411 (2021).
Corces, M. R. et al. The chromatin accessibility landscape of primary human cancers. Science 362, eaav1898 (2018).
Zheng, R. et al. Cistrome Data Browser: expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res. 47, D729–D735 (2019).
Kamimoto, K., Hoffmann, C. M. & Morris, S. A. Dissecting cell identity via network inference and in silico gene perturbation. Nature 614, 742–751 (2023).
Pliner, H. A. et al. Cicero predicts cis-regulatory dna interactions from single-cell chromatin accessibility data. Mol. Cell 71, 858–871 (2018).
Qiu, X. et al. Mapping transcriptomic vector fields of single cells. Cell 185, 690–711 (2022).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Ogishi, M. et al. Multibatch cytometry data integration for optimal immunophenotyping. J. Immunol. 206, 206–213 (2021).
Van Gassen, S. et al. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytom. A 87, 636–645 (2015).
Nowicka, M. et al. CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. F1000Res. 6, 748 (2017).
Diggins, K. E., Greenplate, A. R., Leelatian, N., Wogsland, C. E. & Irish, J. M. Characterizing cell subsets using marker enrichment modeling. Nat. Methods 14, 275–278 (2017).
Street, K. et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018).
Argelaguet, R. et al. MOFA+: a statistical framework for comprehensive integration of multi-modal single-cell data. Genome Biol. 21, 111 (2020).
Newman, A. M. et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 37, 773–782 (2019).
Acknowledgements
We thank the patients and healthy donors who participated for their generous contribution to this study and the Emergency Medicine Research Oxford team and wider clinical staff, in particular A. Brent, A. Novak, A. Kingwill and R. Sayeed and Oxford University NHS Hospital Trusts recruiting hospitals involved in patient recruitment and sample collection. We thank the Oxford Genomics Centre for sequencing support, in particular A. Lee for project management and I. Nassiri for bioinformatic support. We thank the Wellcome Centre for Human Genetics and Jenner Institute flow facilities for assistance in sorting experiments. We are grateful to M. Attar, F. Curion, K. Kedzierska, P. Maclean, G. Scozzafava and members of the Knight laboratory for support and critical discussion. We also thank the COMBAT consortium for access to data for re-analysis.
We acknowledge the support of the National Institute for Health Research (NIHR) through the Comprehensive Clinical Research Network for patient recruitment. This research was funded by the Medical Research Council (MR/V002503/1) (to J.C.K. and E.E.D.), Wellcome Trust Investigator Award (204969/Z/16/Z, to J.C.K.; 209422/Z/17/Z, to I.A.U.), Chinese Academy of Medical Sciences Innovation 537 Fund for Medical Science (2018-I2M-2-002; to J.C.K.), Croucher Foundation (to A.J.K.), Wellcome Trust Grants (090532/Z/09/Z and 203141/Z/16/Z) to core facilities Wellcome Centre for Human Genetics, Wellcome Trust core funding to the Wellcome Sanger Institute (grant numbers WT206194 and 108413/A/15/D) and NIHR Oxford Biomedical Research Centre (to J.C.K.). The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the UK Department of Health. For the purpose of Open Access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission.
Author information
Authors and Affiliations
Consortia
Contributions
Conceptualization was the responsibility of A.J.K. and J.C.K. Data curation was conducted by A.J.K. Formal analysis was conducted by A.J.K., E.C.-G., A.A., M.S. and Y.-X.Z. Funding was acquired by J.C.K. and E.E.D. Investigation was carried out by A.J.K., A.A., M.S. and R.C.F. Methodology was the responsibility of A.J.K., E.C.-G. and K.L.B. Project administration was carried out by J.C.K., C.M., I.A.U., J.A.T. and E.E.D. Resources were the responsibility of A.J.K., E.C.-G., J.C.K., A.J.M., S.M. and C.J.H. Software was the responsibility of A.J.K. and E.C.-G. Supervision was carried out by J.C.K., I.A.U. and C.M. Visualization was carried out by A.J.K. and J.C.K. Writing of the original draft was conducted by A.J.K. and J.C.K. Review and editing was carried out by all authors who read, provided input on and approved the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks John Marshall and Renato Ostuni for their contribution to the peer review of this work. Primary Handling Editor: Ioana Visan, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 scWB single-cell RNA and cell surface profiling annotation, clustering and neutrophil RNA velocity analysis.
a, UMAP of 272,993 cells with TotalVI denoised surface protein marker expression overlaid for selected lineage defining markers. b, Cell surface expression of proteins denoting neutrophil maturation stages for annotated neutrophil subsets. Boxplots denote minimum and maximum with whiskers and bottom quartile, median and upper quartile with the box. c, Comparison of broad annotations (Methods ‘Whole blood single cell multi-omic analysis (scWB)’) with data driven algorithmic labeling of cell identity by SingleR based on reference bulk RNA-seq profiles of pure cell populations from the Blueprint Consortium. d, Proportions of neutrophils and lymphocytes and HLA-DRA and HLA-DRB1 gene expression in classical monocytes in healthy controls (HC) (n = 6), sepsis (n = 26) and post-cardiac surgery (CS) (n = 7) samples. Boxplots denote minimum and maximum with whiskers and bottom quartile, median and upper quartile with the box. e, Clustering results at varying resolutions (0.8–1.1) for fine annotation strategy. f–i, Heat maps of gene markers for (f) PADI4+ immature neutrophils (g) IL1R2+ immature neutrophils (h) MPO+ immature neutrophils/progenitors and (i) cycling neutrophil progenitors. j, UMAP of neutrophils (excluding degranulating neutrophils) with RNA velocity stream directions plotted. k, Partition-based graph abstraction analysis of RNA velocity predicted cellular trajectory transitions. Neu, neutrophils.
Extended Data Fig. 2 ScWB healthy control (HC)-post-cardiac surgery (CS) and CS-sepsis cell type/state differential abundance (DA) analysis and sepsis neutrophil-CD4+ T cell immunosuppression.
a, DA analysis using graph neighborhood-based method to detect enrichment of neighborhoods between CS and HC samples, with UMAP of sampled neighborhoods and statistically significant enrichment colored (spatial FDR < 0.05 with generalized linear modeling). b, Beeswarm plot of CS-HC DA analysis with cluster labels of neighborhoods depicted. c, UMAP of DA analysis between CS and sepsis. Nhood, neighborhood; logFC, log fold change. d, Proportions of immature neutrophil populations as a percentage of total neutrophils across comparator groups (n = 26 sepsis, n = 6 HC, n = 7 CS). Boxplots denote minimum and maximum with whiskers and bottom quartile, median and upper quartile with the box. e, Percentage of live, non-apoptotic CD4+ T cells after 72–96 hours of co-culture with either HC or sepsis neutrophils. Error bars denote standard error of the mean. f, Correlations (Spearman’s Rho) of proportions of each neutrophil subset as measured from single-cell RNA and cell surface protein profiling from sepsis patients with neutrophil-CD4+ T cell co-culture suppression readouts (n = 11). g, Percentage of CD4+ T cells proliferating, expressing PD-1 or expressing CD69 in sepsis neutrophil-allogeneic T cell co-cultures with various treatments compared to untreated (UT) control (n = 6). Untreated (UT), 15 μM TG6−10-1 (PG EP2 receptor antagonist), 15 μM GW-627368 (PG EP4 receptor antagonist), 10 μM indomethacin (COX inhibitor), 10 ng/mL anti-PD-L1/2, 1 mM L-arginine + 1 μM arginase-1 inhibitor. Boxplots denote minimum and maximum with whiskers and bottom quartile, median and upper quartile with the box. h, Gene set enrichment analysis of MsigDB C2 prostaglandin related pathways for sepsis vs. HC neutrophil differential gene expression. i, Violin plots of gene set scoring of mature and immature neutrophils in sepsis and healthy control (HC) samples by gene signatures of granulocytic myeloid-derived suppressor cells (G-MDSC)22,23. P-values for gene set scoring were calculated with two-sided Wilcoxon rank-sum tests while P-values for co-culture inhibition reversal were calculated with two-sided Wilcoxon signed-rank tests. Neu, neutrophils; Cycling progen, cycling neutrophil progenitors; IL1R2+/IL1R2+ immature, IL1R2+ immature neutrophils; MPO+, MPO+ immature neutrophils or progenitors; PADI4+, PADI4+ immature neutrophils; PG, prostaglandin; COX, cyclo-oxygenase; NES, normalized enrichment score; FDR, false discovery rate. *P < 0.05, ****P < 0.0001.
Extended Data Fig. 3 scWB convalescent sepsis neutrophil analysis and scHSPC hematopoietic stem and progenitor cell (HSPC) RNA based identity mapping.
a, Boxplots of proportions of neutrophil states in HC vs. convalescent sepsis samples from scWB. Boxplots denote minimum and maximum with whiskers and bottom quartile, median and upper quartile with the box. b, Proliferative fraction of anti-CD3/28 bead-stimulated CD4+ T cells after co-culture with neutrophils at a 4 neutrophil:1 T cell ratio, compared to positive controls of CD4+ T cells cultured with anti-CD3/28 beads alone (n = 10 HC, n = 9 convalescent sepsis). Boxplots denote minimum and maximum with whiskers and bottom quartile, median and upper quartile with the box. c,d, HSPC (right) mapping to (c) Seurat bone marrow mononuclear cell (BMMC) dataset24 (left) and (d) healthy donor BMMC dataset from Granja et al.25 (left). e, UMAP of HSPCs clustered on Harmony batch corrected RNA and chromatin reduced dimensions after non-HSPCs were filtered out (46,156 cells; Methods). f, UMAP colored by RNA prediction identities from HSPC mapping to Seurat BMMC dataset. g, UMAP of progenitor cells alone clustered on Harmony batch corrected RNA and chromatin reduced dimensions (16,820 cells; Methods). h, Boxplots of progenitor cell clusters as a proportion of total progenitor cells (n = 15 acute sepsis, n = 7 HC). Boxplots denote minimum and maximum with whiskers and bottom quartile, median and upper quartile with the box. i, Heat map of genes defining progenitor clusters with known lineage defining genes highlighted. j, Boxplots of HSC clusters as a proportion of total HSCs (n = 15 acute sepsis, n = 7 HC). Boxplots denote minimum and maximum with whiskers and bottom quartile, median and upper quartile with the box. k, UMAP of CD34+ HSPCs from annotated Human Cell Atlas bone marrow mononuclear cell (BMMC) scRNA-seq data26 for deriving granulopoiesis gene set (Methods). Neu, neutrophils; c/pDC, conventional/plasmacytoid dendritic cell; GMP, granulocyte monocyte progenitor; LMPP, lymphoid-primed multipotential progenitors; NK, natural killer; Prog, progenitor; Mk, megakaryocyte; RBC, red blood cell; Eryth, erythrocyte; CMP, common myeloid progenitor; Mono, monocyte; CD4 N1/2, CD4+ naïve T cell 1/2; CD4 M, memory CD4+ T cell; CD8 CM, central memory CD8+ T cell; PC, plasma cell; MDP, monocyte/dendritic cell progenitor; MultiLin, multilineage progenitor; MEP, megakaryocyte/erythroid progenitor; MKP megakaryocyte progenitor; ERP, erythrocyte progenitor; Eo, eosinophil; Mast, mast cell.
Extended Data Fig. 4 scWB scRNA-seq data substructure and concordance with transcriptomic sepsis response signature (SRS) endotypes.
a, First two principal components from principal components analysis of pseudobulked single-cell expression profiles of sepsis samples colored by SRS assignment. b, Pearson’s correlation of fold change in gene expression between SRS1 vs. non-SRS1 (sample level pseudobulks) in scWB and cohort in which SRS was originally derived5. c, Consensus clustering of pseudobulked single-cell gene expression profiles of sepsis samples (all cells per sample) at the level of two clusters. d, Overlaps of consensus clustering classification vs. SRS classification. e, Unsupervised hierarchical clustering of pseudobulked single-cell expression profiles of sepsis samples. SRS, sepsis response signature.
Extended Data Fig. 5 scWB scRNA-seq SRS gene expression analysis and SRS neutrophil functional interrogation.
a, b, Volcano plots of DGE analysis between SRS groups for pseudobulked (a) neutrophil states and (b) mononuclear cells (red denoting genes with fold change > 1.2 and FDR < 0.1) (positive fold change denoting upregulation in SRS1). c, Correlation (Spearman’s Rho 95% confidence interval) of SRS1 sample median neutrophil IL1R2 gene expression as measured by scRNA-seq with median neutrophil IL1R2 cell surface protein expression as measured by flow cytometry (n = 14). d, Proliferative fraction of anti-CD3/28 bead-stimulated CD4+ T cells after co-culture with neutrophils at a 4 neutrophil:1 T cell ratio, compared to positive controls of CD4+ T cells cultured with anti-CD3/28 beads alone (n = 6 SRS1, n = 13 non-SRS1). Boxplots denote minimum and maximum with whiskers and bottom quartile, median and upper quartile with the box. e, Neutrophil phagocytosis with and without GM-CSF stimulation (n = 15). Boxplots denote minimum and maximum with whiskers and bottom quartile, median and upper quartile with the box. P-values for neutrophil functional assays were calculated with two-sided Wilcoxon rank-sum tests. SRS, sepsis response signature; FDR, false discovery rate; ns, not significant. Neu, neutrophils.
Extended Data Fig. 6 MmV validation of neutrophil immaturity underlying SRS1.
a, First two principal components from principal components analysis of n = 42 sepsis patient samples (36 individuals) for peripheral blood bulk RNA-seq (23,063 genes). b, Kaplan-Meier survival by SRS (log-rank test P-value, 95% confidence intervals). c, Heat map of 15 immune cell clusters (with neutrophil clusters merged; Methods) from clustering on 41 protein markers for n = 41 sepsis (36 individuals) and n = 11 healthy control (HC) samples. d, UMAP of nine broad immune cell types (2,287,410 cells) from CyTOF immunophenotyping. e, Marker enrichment modeling (MEM) score heat map of eight neutrophil clusters (1,921,471 cells), identified from neutrophil subclustering on 17 selected protein markers. f, g, UMAP of (f) neutrophil subsets showing (g) density-based distribution by SRS/healthy control (HC) status. h, Neutrophil cluster frequency as a proportion of all cells with proportional differences across SRS analyzed by generalized linear mixed models (n = 41 samples, 36 individuals). i, Diffusion map dimensionality reduction of neutrophils. Boxplots denote minimum and maximum with whiskers and bottom quartile, median and upper quartile with the box. j, Diffusion components 1 and 2 with black line depicting trajectory identified by principal curve fitting. Color reflects pseudotime value of cell with red indicating early pseudotime and blue late pseudotime. k, Dotplot of neutrophil subsets according to pseudotemporal ordering from principal curve trajectory with median and interquartile range. l, Density distribution of neutrophils over pseudotime with Kolmogorov-Smirnov test for SRS1 vs. non-SRS1 distributions. m, CyTOF PCA using proportions of all cells (left), only mononuclear cells (MNC, middle) and only neutrophils (right) with 95% data ellipses (assuming a multivariate t-distribution). SRS, sepsis response signature; cMono, classical monocyte; ncMono, non-classical monocyte; NK cell, natural killer cell; Neu, neutrophil; pro-neu, pro-neutrophil; pre-neu, pre-neutrophil; imm neu, immature neutrophil; int neu, intermediate neutrophil; int inf neu, intermediate inflammatory neutrophil; mature neu, mature neutrophil; MNP, mononuclear phagocytes; DC, diffusion component; FDR, false discovery rate. * FDR < 0.05, ** FDR < 0.01, *** FDR < 0.001.
Extended Data Fig. 7 MmV factor analysis (MOFA+) and brWB-CID validation of SRS1 IL1R2+ immature neutrophil expansion.
a-j MmV. a, Differentially expressed modules in whole blood bulk RNA-seq (FDR < 0.01, two-sided Wilcoxon rank-sum test) (n = 26 SRS1, n = 16 non-SRS1). Violin plots with median, 95% confidence interval and interquartile range. b, Enrichment of neutrophil related gene sets of weighted gene co-expression network analysis (WGCNA) modules upregulated in SRS1. c, Correlation (and 95% confidence interval) of module 10 eigengene expression with percentage of immature neutrophils from mass cytometry (CyTOF) per sample of mmV. d, Leading edge analysis for enrichment of scWB derived IL1R2+ immature neutrophil defining signature in module 10. e, IL1R2+ immature neutrophil (top) and cycling neutrophil progenitor (bottom) proportions following total leukocyte bulk transcriptomics deconvolution of n = 542 sepsis patients (brWB-CID microarray data)5,18. Boxplots denote minimum and maximum with whiskers and bottom quartile, median and upper quartile with the box. f, Input data into MOFA+77 model consisting of three modalities (105 plasma proteins, 33 module eigengenes (eigen) from WGCNA and 22 cell clusters from CyTOF) for n = 36 sepsis samples with one and six missing samples for CyTOF and plasma proteomics respectively. g, Scatter-plots of latent factor values for sepsis patients. h, Factor values by SRS groups (two-sided Wilcoxon rank-sum test) (n = 26 SRS1, n = 16 non-SRS1). Violin plots with median, 95% confidence interval and interquartile range. i, Variance decomposition showing percentage variance explained for individual modalities by each latent factor. j, Latent factor two weights per cell cluster (top) and gene module (bottom). k, Latent factor one weights per cell cluster (top) and gene module (bottom). SRS, sepsis response signature; Neu, neutrophil; imm neu, immature neutrophil; int inf neu, intermediate inflammatory neutrophil; pro-neu, pro-neutrophil; pre neu, pre-neutrophil; cMono, classical monocyte; ncMono non-classical monocyte; intMono, intermediate monocyte; cMono_1; Ki67+, cMono_2: CD163+CD354+Ki67+, cMono_3: CD163+, cMono_4: CD354+Ki67+. NES, normalized enrichment score; FDR, false discovery rate. * FDR < 0.05, ** FDR < 0.01, **** FDR < 0.0001, NS, not significant.
Extended Data Fig. 8 Hematopoietic stem cell (HSC) SRS differential abundance (DA) and clusters C5 and C7 peak set analysis.
a, HSC DA across SRS with UMAP showing sampled neighborhoods colored by statistically significant enrichment (spatial FDR < 0.05). b, c, Volcano plots of differentially accessible (DA) peaks (fold change > 1.5, FDR < 0.05) for HSC (b) cluster C5 vs. all other clusters and (c) cluster C7 s. all other clusters (Wilcoxon rank-sum test). d, e, Enrichment of bulk ATAC-seq profiles in differentially open peaks for clusters (d) C5 (184 peaks) and (e) C7 (718 peaks). f, g, Enrichment of transcription factor (TF) motifs of differentially open peaks for clusters (f) C5 and (g) C7. h, ChIP-seq overlap analysis for cluster C7 differentially open peaks. The Giggle score denotes a composite significance and effect size scoring (doi:10.1038/nmeth.4556). Nhood, neighborhood; SRS, sepsis response signature; FDR, false discovery rate; ChIP, chromatin immunoprecipitation.
Extended Data Fig. 9 Hematopoietic stem cell (HSC) trajectory and vector field analysis, and transcription factor (TF) knockout and overexpression analysis.
a, UMAP of HSCs with supervised pseudotime trajectory from cluster C6 (HC enriched) to cluster C7 (sepsis but not SRS1 enriched). b, STAT3 and CEBPB gene expression with pseudotime. c, Genes (left) and TF motifs (right) which change along C6−C7 pseudotime trajectory and correlate with each other (Pearson’s r > 0.5, FDR < 0.05). d, CEBPA gene expression with pseudotime. e, CellOracle68 in silico knockout (top) and overexpression (bottom) of CEBPA and CEBPB and effects on clusters C5/6/7, with arrows displaying predicted changes in cell fate after gene of interest is perturbed (Methods). f–i, HSC clusters C5/6/7 vector field analysis with dynamo70. f, UMAP of HSC clusters C5/6/7 with RNA velocity stream directions plotted. g, RNA velocity based pseudotime of HSC clusters C5/6/7. h, dynamo acceleration and curvature vector fields. i, In silico knockout of CEBPA (left), CEBPB (middle) and STAT3 (right) and effects on clusters C5/6/7 with arrows displaying predicted changes in cell fate after gene of interest is perturbed. j, dynamo Jacobian analysis of gene-gene regulatory relationships. Scatter-plots show normalized expression values of genes of interest along x and y axes. Blue denotes inhibition of the numerator gene by the denominator gene in the Jacobian partial derivative, while red denotes activation of the numerator by the denominator.
Extended Data Fig. 10 SRS1 signatures across differing clinical contexts of infectious disease.
a–e, BrWB-CID contexts 2–7. a, Cycling neutrophil progenitor proportions following whole blood bulk transcriptomics deconvolution in 6 cohorts of infectious disease patients defined by infecting organism (n = 77 COVID-19 and n = 109 influenza, top), source of infection (n = 438 community acquired pneumonia and n = 229 fecal peritonitis, middle) and clinical syndrome (n = 77 adult acute respiratory distress syndrome (ARDS) and n = 106 pediatric septic shock, bottom). Boxplots denote minimum and maximum with whiskers and bottom quartile, median and upper quartile with the box. Two-sided Wilcoxon rank-sum test comparing SRS1 and non-SRS1 groups. b, Gene set enrichment analysis of scWB STAT3 gene expression programs (GEPs) (the union of all three GEPs, STAT3-union-GEP) in SRS1 upregulated genes for each of the 6 cohorts. c, IL1R2+ immature neutrophil proportions in COVID-19 whole blood scRNA-seq cohort (n = 8) after referencing mapping of dataset to scWB reference single cell atlas. d, Violin plots of scWB neutrophil STAT3 GEP expression in COVID-19 neutrophils as defined by scWB neutrophil states. Two-sided Wilcoxon rank-sum test comparing SRS1 and non-SRS1 groups. e, Violin plots of scWB neutrophil STAT3-union-GEP expression in all COVID-19 neutrophils. Two-sided Wilcoxon rank-sum test comparing SRS1 and non-SRS1 groups. SRS, sepsis response signature; Neu, neutrophil. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Supplementary information
Supplementary Information
Supplementary Note 1.
Supplementary Table 1
Clinical data for cohorts 1 and 3.
Supplementary Table 2
Antibodies used for cell surface protein profiling for scWB on BD Rhapsody platform.
Supplementary Table 3
Gene markers for each annotated cell type/state in scWB single-cell profiling and top 50 genes of each program for neutrophil states from scWB neutrophil consensus non-negative matrix factorization.
Supplementary Table 4
MsigDB Hallmark pathways and prostaglandin gene sets enriched from scWB neutrophil consensus non-negative matrix factorization.
Supplementary Table 5
Differentially accessible peaks (fold change > 1.5, FDR < 0.05) for HSC clusters C5 and C7.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kwok, A.J., Allcock, A., Ferreira, R.C. et al. Neutrophils and emergency granulopoiesis drive immune suppression and an extreme response endotype during sepsis. Nat Immunol 24, 767–779 (2023). https://doi.org/10.1038/s41590-023-01490-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-023-01490-5
This article is cited by
-
Monocyte state 1 (MS1) cells in critically ill patients with sepsis or non-infectious conditions: association with disease course and host response
Critical Care (2024)
-
Monocytic myeloid-derived suppressor cells as an immune indicator of early diagnosis and prognosis in patients with sepsis
BMC Infectious Diseases (2024)
-
The pathophysiology of sepsis and precision-medicine-based immunotherapy
Nature Immunology (2024)
-
Integrating single-cell multi-omics and prior biological knowledge for a functional characterization of the immune system
Nature Immunology (2024)
-
CD200Rhigh neutrophils with dysfunctional autophagy establish systemic immunosuppression by increasing regulatory T cells
Cellular & Molecular Immunology (2024)