Abstract
The high mobility group (HMG) transcription factor TCF-1 is essential for early T cell development. Although in vitro biochemical assays suggest that HMG proteins can serve as architectural elements in the assembly of higher-order nuclear organization, the contribution of TCF-1 on the control of three-dimensional (3D) genome structures during T cell development remains unknown. Here, we investigated the role of TCF-1 in 3D genome reconfiguration. Using gain- and loss-of-function experiments, we discovered that the co-occupancy of TCF-1 and the architectural protein CTCF altered the structure of topologically associating domains in T cell progenitors, leading to interactions between previously insulated regulatory elements and target genes at late stages of T cell development. The TCF-1-dependent gain in long-range interactions was linked to deposition of active enhancer mark H3K27ac and recruitment of the cohesin-loading factor NIPBL at active enhancers. These data indicate that TCF-1 has a role in controlling global genome organization during T cell development.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Genomics data generated in this study are publicly available in the Gene Expression Omnibus at accession code GSE178348. Oligopaint FISH data generated in this study are provided as Source Data. Source data are provided with this paper.
Code availability
The codes for Stripenn and data analysis are available from the author’s GitHub page (https://github.com/VahediLab/TCF13D_code).
References
Beagan, J. A. & Phillips-Cremins, J. E. On the existence and functionality of topologically associating domains. Nat. Genet. 52, 8–16 (2020).
Dixon, J. R. et al. Chromatin architecture reorganization during stem cell differentiation. Nature 518, 331–336 (2015).
Dowen, J. M. et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 159, 374–387 (2014).
Hnisz, D., Shrinivas, K., Young, R. A., Chakraborty, A. K. & Sharp, P. A. A phase separation model for transcriptional control. Cell 169, 13–23 (2017).
Nora, E. P. et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Hansen, A. S. CTCF as a boundary factor for cohesin-mediated loop extrusion: evidence for a multi-step mechanism. Nucleus 11, 132–148 (2020).
Stadhouders, R., Filion, G. J. & Graf, T. Transcription factors and 3D genome conformation in cell-fate decisions. Nature 569, 345–354 (2019).
Stadhouders, R. et al. Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat. Genet. 50, 238–249 (2018).
Weintraub, A. S. et al. YY1 is a structural regulator of enhancer-promoter loops. Cell 171, 1573–1588 (2017).
Di Giammartino, D. C. et al. KLF4 is involved in the organization and regulation of pluripotency-associated three-dimensional enhancer networks. Nat. Cell Biol. 21, 1179–1190 (2019).
Johanson, T. M. et al. Transcription-factor-mediated supervision of global genome architecture maintains B cell identity. Nat. Immunol. 19, 1257–1264 (2018).
Singh, H., Khan, A. A. & Dinner, A. R. Gene regulatory networks in the immune system. Trends Immunol. 35, 211–218 (2014).
Allman, D. et al. Thymopoiesis independent of common lymphoid progenitors. Nat. Immunol. 4, 168–174 (2003).
Li, L., Leid, M. & Rothenberg, E. V. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science 329, 89–93 (2010).
Kueh, H. Y. et al. Asynchronous combinatorial action of four regulatory factors activates Bcl11b for T cell commitment. Nat. Immunol. 17, 956–965 (2016).
Johnson, J. L. et al. Lineage-determining transcription factor TCF-1 initiates the epigenetic identity of T cells. Immunity 48, 243–257 (2018).
Emmanuel, A. O. et al. TCF-1 and HEB cooperate to establish the epigenetic and transcription profiles of CD4(+)CD8(+) thymocytes. Nat. Immunol. 19, 1366–1378 (2018).
Giese, K., Cox, J. & Grosschedl, R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell 69, 185–195 (1992).
Love, J. J. et al. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature 376, 791–795 (1995).
Shan, Q. et al. Tcf1 and Lef1 provide constant supervision to mature CD8(+) T cell identity and function by organizing genomic architecture. Nat. Commun. 12, 5863 (2021).
Zhou, Y. et al. EBF1 nuclear repositioning instructs chromatin refolding to promote therapy resistance in T leukemic cells. Mol. Cell 82, 1003–1020 e1015 (2022).
Antoszewski, M. et al. Tcf1 is essential for initiation of oncogenic Notch1-driven chromatin topology in T-ALL. Blood 139, 2483–2498 (2022).
Dose, M. et al. β-Catenin induces T-cell transformation by promoting genomic instability. Proc. Natl Acad. Sci. USA 111, 391–396 (2014).
Hu, G. et al. Transformation of accessible chromatin and 3D nucleome underlies lineage commitment of early T cells. Immunity 48, 227–242 e228 (2018).
Crane, E. et al. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523, 240–244 (2015).
Fasolino, M. et al. Genetic variation in type 1 diabetes reconfigures the 3D chromatin organization of T cells and alters gene expression. Immunity 52, 257–274 (2020).
Kernfeld, E. M. et al. A single-cell transcriptomic atlas of thymus organogenesis resolves cell types and developmental maturation. Immunity 48, 1258–1270 (2018).
Zhang, J. A., Mortazavi, A., Williams, B. A., Wold, B. J. & Rothenberg, E. V. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell 149, 467–482 (2012).
Kloetgen, A. et al. Three-dimensional chromatin landscapes in T cell acute lymphoblastic leukemia. Nat. Genet. 52, 388–400 (2020).
Dekker, J. et al. The 4D nucleome project. Nature 549, 219–226 (2017).
Krijger, P. H. et al. Cell-of-origin-specific 3D genome structure acquired during somatic cell reprogramming. Cell Stem Cell 18, 597–610 (2016).
Dionne, C. J. et al. Subversion of T lineage commitment by PU.1 in a clonal cell line system. Dev. Biol. 280, 448–466 (2005).
Forcato, M. et al. Comparison of computational methods for Hi-C data analysis. Nat. Methods 14, 679–685 (2017).
Eres, I. E. & Gilad, Y. A TAD skeptic: is 3D genome topology conserved? Trends Genet. 37, 216–223 (2021).
Beliveau, B. J. et al. Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat. Commun. 6, 7147 (2015).
Beliveau, B. J. et al. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc. Natl Acad. Sci. USA 109, 21301–21306 (2012).
Ollion, J., Cochennec, J., Loll, F., Escude, C. & Boudier, T. TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics 29, 1840–1841 (2013).
Ciosk, R. et al. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell 5, 243–254 (2000).
Murayama, Y. & Uhlmann, F. Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature 505, 367–371 (2014).
Petela, N. J. et al. Scc2 is a potent activator of cohesin’s ATPase that promotes loading by binding Scc1 without Pds5. Mol. Cell 70, 1134–1148 e1137 (2018).
Vian, L. et al. The energetics and physiological impact of cohesin extrusion. Cell 173, 1165–1178 (2018).
Fudenberg, G. et al. Formation of chromosomal domains by loop extrusion. Cell Rep. 15, 2038–2049 (2016).
Kueng, S. et al. Wapl controls the dynamic association of cohesin with chromatin. Cell 127, 955–967 (2006).
Rao, S. S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Zhu, Y., Denholtz, M., Lu, H. & Murre, C. Calcium signaling instructs NIPBL recruitment at active enhancers and promoters via distinct mechanisms to reconstruct genome compartmentalization. Genes Dev. 35, 65–81 (2021).
Beagan, J. A. et al. Three-dimensional genome restructuring across timescales of activity-induced neuronal gene expression. Nat. Neurosci. 23, 707–717 (2020).
Petrovic, J. et al. Oncogenic notch promotes long-range regulatory interactions within hyperconnected 3D Cliques. Mol. Cell 73, 1174–1190 (2019).
Yoshida, H. et al. The cis-regulatory atlas of the mouse immune system. Cell 176, 897–912 (2019).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Servant, N. et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16, 259 (2015).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Durand, N. C. et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98 (2016).
Wolff, J. et al. Galaxy HiCExplorer: a web server for reproducible Hi-C data analysis, quality control and visualization. Nucleic Acids Res. 46, W11–W16 (2018).
Conway, J. R., Lex, A. & Gehlenborg, N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940 (2017).
Ramirez, F., Dundar, F., Diehl, S., Gruning, B. A. & Manke, T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014).
Lareau, C. A. & Aryee, M. J. hichipper: a preprocessing pipeline for calling DNA loops from HiChIP data. Nat. Methods 15, 155–156 (2018).
Flyamer, I. M., Illingworth, R. S. & Bickmore, W. A. Coolpup.py: versatile pile-up analysis of Hi-C data. Bioinformatics 36, 2980–2985 (2020).
Carleton, M. et al. Signals transduced by CD3ε, but not by surface pre-TCR complexes, are able to induce maturation of an early thymic lymphoma in vitro. J. Immunol. 163, 2576–2585 (1999).
de Boer, J. et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 33, 314–325 (2003).
Ogilvy, S. et al. Promoter elements of vav drive transgene expression in vivo throughout the hematopoietic compartment. Blood 94, 1855–1863 (1999).
Shimshek, D. R. et al. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis 32, 19–26 (2002).
Yang, Q. et al. TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nat. Immunol. 16, 1044–1050 (2015).
Chen, Z. et al. In vivo CD8(+) T cell CRISPR screening reveals control by Fli1 in infection and cancer. Cell 184, 1262–1280 (2021).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Mumbach, M. R. et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 13, 919–922 (2016).
Beliveau, B. J. et al. OligoMiner provides a rapid, flexible environment for the design of genome-scale oligonucleotide in situ hybridization probes. Proc. Natl Acad. Sci. USA 115, E2183–E2192 (2018).
Moffitt, J. R. & Zhuang, X. RNA imaging with multiplexed error-robust fluorescence in situ hybridization (MERFISH). Methods Enzymol. 572, 1–49 (2016).
Rosin, L. F., Nguyen, S. C. & Joyce, E. F. Condensin II drives large-scale folding and spatial partitioning of interphase chromosomes in Drosophila nuclei. PLoS Genet. 14, e1007393 (2018).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Yoon, S., Chandra, A. and Vahedi, G. Stripenn detects architectural stripes from chromatin conformation data using computer vision. Nat. Commun. https://doi.org/10.1038/s41467-022-29258-9 (2022).
Abdennur, N. & Mirny, L. A. Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics 36, 311–316 (2020).
Imakaev, M. et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat. Methods 9, 999 (2012).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Acknowledgements
We thank R. B. Faryabi, K. Zaret, A. Karnay, E. J. Wherry and R. Jain for helpful discussions. We thank J. Shi and his laboratory for help and discussion about the CRISPR/Cas9 plasmids. This work was supported by National Institutes of Health grants UC4 DK112217, U01DK112217, R01HL145754, U01DK127768 and U01DA052715, the Burroughs Wellcome Fund, the Chan Zuckerberg Initiative, W. W. Smith Charitable Trust, the Penn Epigenetics Institute pilot and the Sloan Foundation awards to G.V.
Author information
Authors and Affiliations
Contributions
A.C. performed all genomics and sequencing experiments in collaboration with N.G. N.G. generated the Tcf7−/− mice. A.C. performed computational analysis of ChIP-seq data in DN3. W.W. performed all computational analysis of genomics data in the first submission and wrote the initial draft of the first submission. S.Y. performed all computational analysis for genomics and imaging data for two revisions. E.K.F. performed all FISH experiments and assisted in mouse breeding. S.C.N and E.F.J. assisted in Oligopaint probe design. G.V. conceptualized and supervised the project, wrote the manuscript, revised it with comments from all authors and acquired funding.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks the anonymous reviewers for their contribution to the peer review of this work. Ioana Visan was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Loops associated with CTCF and TCF-1.
Genome browser view showing cohesin loops and CTCF, TCF-1, SMC1 binding at the Ets1 (a) and Tcf7 (b) loci. The loops associated with both TCF-1 and CTCF are shown in the bottom panel.
Extended Data Fig. 2 Loss of insulation and boundaries during T cell development.
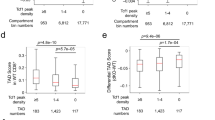
a. Summary plot and heat map of insulation score in six T cell developmental stages at CTCF only (red), CTCF+TCF-1 (yellow), TCF-1-only (blue) peaks and random regions as control (black), where lower value indicates higher insulation. Peaks defined in Fig. 1c. b. Violin plot and boxplot (center, median; box limits, upper (75th) and lower (25th) percentiles; whiskers, 1.5× interquartile range; points, outliers) showing differences in insulation score from CLP to DP at the four different sets of peaks. The comparison between different groups was done with one-sided Student’s t-test. The insulation score change was significantly (P < 2.2 × 10−16) different between different sets of peaks. c. Bar plot showing the total number of boundaries in six T cell developmental stages. d. Upset plot showing unique and shared boundaries among the six T cell developmental stages. The color shows different groups of boundaries that are lost or gained during T cell development. e. Local pileup in six T cell developmental stages at the nine groups of boundaries classified in (d). f. Quantification of interactions in six T cell developmental stages between upstream and downstream regions of the nine groups of boundaries.
Extended Data Fig. 3 Chromatin features at dynamic boundaries.
a. Summary plot and heat map of insulation score in 1 Mbp regions centered at the nine groups of boundaries in six T cell developmental stages. b. Summary plot and heat map of directionality index at the nine groups of boundaries in six T cell developmental stages. Cumulative distribution of PC1 values in CLP cells (c) and DP cells (d) at nine boundary groups. Cumulative distribution of CTCF only peak density (e), CTCF+TCF-1 peaks density (f) and TCF-1-only peak density (g) as defined in Fig. 1c at nine boundary groups h. Heatmap showing enrichment of CTCF only, CTCF+TCF-1, TCF-1 only peaks and random regions at nine boundary groups, as well as the H3K4me2, H3KAc, ATAC-seq and RNA-seq signals at T cell developmental stages. Signals in each row were normalized between 0 and 1. i. Heat map showing the statistical test of enrichment of epigenetic features at each group of boundaries compared with conserved boundaries using Kolmogorov–Smirnov test. j. Genome browser view and contact matrix at the Rag1/2 locus. Contact matrix was normalized according to the sequencing depth and triangles show the identified TADs in this region. Insulation score depicted as a line plot and colored according to their developmental stage as indicated in the figure legend. Genes in this region are shown with Rag1 and Rag2 genes highlighted in orange.
Extended Data Fig. 4 TCF-1 and CTCF co-bound sites are evolutionarily conserved and insulated in non-T cells.
a. Summary plot and heat map of sequence conservation using phastCons score at CTCF only, CTCF+TCF-1, TCF-1-only peaks and random regions as control. b. Local pileup plot of interactions using Hi-C measurements in progenitor and mature T cells in humans at homologs CTCF and TCF-1 peaks. Hi-C measurements were from human publicly available T-ALL, ETP T-ALL and ultra-deep Hi-C in CD8+ T cells from a healthy donor generated for this study. Human homologous regions for CTCF only, TCF-1 only as well as TCF-1+CTCF peaks in mice were found using liftOver. c. Local pileup plot of interactions at CTCF only, TCF-1+CTCF, TCF-1 only peaks in different mouse cell types, including mouse embryonic stem cell (mESC), fibroblast (NIH 3T3), cardiomyocyte, B cells, naïve CD4+ and DP T cells. d. Local pileup plot of interactions in different human cell types from 4D Nucleome repository at the liftOver sites of CTCF only, TCF-1 only as well as CTCF+TCF-1 peaks from mice.
Extended Data Fig. 5 Differential gene expression and CTCF binding after TCF-1 overexpression in fibroblast.
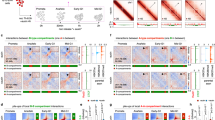
a. Volcano plot of differentially expressed genes in non-induced 3T3 and 3T3TCF-1 fibroblasts. Log2 fold change and P values are calculated with DEseq2. P depicts adjusted pvalue calculated by DESeq2. b. Volcano plot of differential CTCF binding in non-induced 3T3 and 3T3TCF-1 fibroblasts. Log2 fold change and P values are calculated with DEseq2. P depicts adjusted pvalue calculated by DESeq2. c. Scatter plot of PC1 values in non-induced 3T3 and 3T3TCF-1 fibroblasts. The blue lines are where PC1 values are equal to zero and the red line is where PC1 values are equal in the two conditions. Pearson correlation and the two-sided significance level are shown, the PC1 values are significantly (P < 2.2 × 10−16) correlated. d. Saddle plot showing the compartmentalization in non-induced 3T3 and 3T3TCF-1 fibroblasts. The regions were sorted by PC1 value from B to A compartment and interactions between different regions are shown in the heat map. e. Boxplot (center, median; box limits, upper (75th) and lower (25th) percentiles; whiskers, 1.5× interquartile range; points, outliers) shows the compartment strength, BB interactions, AA interactions and AB interactions in wild-type and Tcf7−/− DN3s on all chromosomes (n=20). The statistical test was done using two-sided Student’s t-test. (ns: not significant). f. Venn diagram showing the unique and shared boundaries non-induced 3T3 and 3T3TCF-1 fibroblasts. g. Summary plot and heat map of TCF-1 ChIP-seq performed in NIH 3T3 cells at unique and common boundaries between and after TCF-1 induction. The three different sets of boundaries were normalized to the same height. h. Boxplots (center, median; box limits, upper (75th) and lower (25th) percentiles; whiskers, 1.5× interquartile range; points, outliers) showing the correlation of TCF-1 binding density in NIH 3T3 cells and increase in domain score after TCF-1 overexpression. TADs (n=2573) are grouped based on the increase of domain score and the boxplots show TCF-1 peak density in each group. R and P values are calculated with two-sided Pearson correlation. The results shows TCF-1 binding is significantly (P < 2.2 × 10−16) associated with domain score increase. i. Illustration of our computational strategy to classify TCF-1-dependent 3D genome interactions originating from TCF-1 binding. Genomic regions were divided into bins and classified based on the number of TCF-1 peaks within 50 kb, as well as TCF-1 binding status in the bin. For every genomic bin, we therefore calculated the number of TCF-1 peaks in the 50 kbp neighborhood of the bin and additionally classified these bins into TCF-1 bound and unbound regions. j. Boxplots (center, median; box limits, upper (75th) and lower (25th) percentiles; whiskers, 1.5× interquartile range; points, outliers) show correlation of interactions at bins with different number of TCF-1 binding events within their 50 kb neighborhood. Each bin itself was further divided based on TCF-1 occupancy. k. The greatest increase in 3D interactions after TCF-1 expression in fibroblasts occurs when TCF-1 occupies both anchors and has high density at anchors’ neighboring regions. We calculated the extent of increase in interactions between 10kbp bin pairs which were grouped based on the status of TCF-1 binding and the number of TCF-1 binding events in their 50kbp neighborhood. l. Violin plot and boxplot (center, median; box limits, upper (75th) and lower (25th) percentiles; whiskers, 1.5× interquartile range; points, outliers) showing the number of loops after TCF-1 overexpression in 3T3 at anchors that are bound by CTCF only, CTCF+TCF-1, TCF-1 only and random regions. The statistical test was done with two-sided Student’s t-test. The results show that the gain of loops after TCF-1 overexpression are significantly (P < 2.2 × 10−16) different between different sets of peaks.
Extended Data Fig. 6 3D genome reorganization after TCF-1 deletion in DN3s and DPs.
a. Western blot of TCF-1 in DN3s in which TCF-1 was disrupted using CRISPR/Cas9. Single cell clones in bold indicate those utilized for further experiments. Experiments were carried out twice and similar results were obtained. b. Illustration of breeding strategy for disruption of Tcf7 in mice by crossing Vav-Cre with Tcf7 floxed mice, generating mice in which TCF-1 is conditionally ablated in all hematopoietic cells. c-d. Western blot of TCF-1 in thymocytes from homozygous fl/fl cre+ (Tcf7−/−), heterozygous fl/+ cre+ (N=2) and cre+ mice. (c) Flow cytometric analysis of thymocytes from Vav cre+ control and Tcf7−/− experimental mice. Top panel shows CD4+ and CD8+ populations and is pre-gated on lymphocytes, live and single cells. Bottom panel shows DN populations DN1-DN4 (upper left to lower left quadrant clockwise) as measured based on CD44 and CD25 expression and is pre-gated on lymphocytes, live, single cells and lineage negative cells (d). e-f. Scatter plot showing A/B compartment distribution using correlation of PC1 values between wildtype and Tcf7−/− DN3s (e) and DPs (f). Blue lines are PC1 equals zero and red line is where PC1 are equal in two conditions. Pearson correlation coefficients and P-values are shown. g. Saddle plot shows the compartmentalization in wildtype and Tcf7−/− DN3s. The regions were sorted by PC1 value from B to A compartment and interactions between different regions are shown in the heat map. One Hi-C experiment was performed in the wildtype condition and two Hi-C experiments were performed in two distinct Tcf7−/− DN3 clones. h. Data are shown as boxplots (center, median; box limits, upper (75th) and lower (25th) percentiles; whiskers, 1.5× interquartile range; points, outliers) showing the compartment strength, BB interactions, AA interactions and AB interactions in wildtype and Tcf7−/− DN3s (n=20 mouse chromosomes). The statistical test was done using two-sided Student’s t-test. (ns: not significant, *: P< 0.05). i-j. Venn diagram showing the unique and shared boundaries between wildtype and Tcf7−/− DN3s (i) and DP (j). k-l. Scatter plot showing changes in gene expression located in the top 50 TADs that lost domain score in DN3 (k) and DP (l) T cells. Genes that were significantly (P < 0.05, abs(log2FoldChange) > 0.5) differentially expressed were shown in red. m. Gene-ontology analysis of genes in the top 50 TADs that gained domain score and the top 50 TADs that lost domain score after TCF-1 deletion in DN3s using metaScape. T cell-specific ontology terms are marked in red.
Extended Data Fig. 7 Reorganization of interferon genes by TCF-1.
a,b,d,e. Contact matrix and genome browser views of ultra-deep Hi-C measurements at interferon-induced gene cluster (genes marked in orange) using two independent clones for deleting TCF-1 using CRISPR/Cas9 in DN3 (a), comparing SMC1 HiChIP in DPs of C57BL/6 and NOD mice (b). We used Stripenn to quantify stripiness of the stripe originating from the TCF1 cluster, which showed that NOD has a stripe score of 2.14, while C57BL/6 is -2.3. SMC1 HiChIP in TCF-1 induction in fibroblasts (d) and ultra-deep Hi-C in wildtype and Tcf7−/− DPs (e). The browser view focuses on a TAD that lose intra-TAD interactions after TCF-1 deletion in both replicates, which also have a cluster of TCF-1 binding (gray highlighted) in wildtype DN3s. The CTCF and SMC1 binding in both conditions are also shown. c. Heat map showing the row normalized gene expression level from DPs in NOD and C57BL/6 of the interferon induced genes in the TAD with different intra-TAD interactions in C57BL/6 and NOD mice. Orange bars represent genomic coordinates of interferon genes.
Extended Data Fig. 8 TCF-1 ChIP-seq comparison in DPs and DN3s.
a. Venn diagram of overlapping and unique bound regions by TCF-1 ChIP-seq thymocytes, DN3s and 3T3TCF-1. b. Local pileup plot of long-range interactions using ultra-deep Hi-C in wildtype and two Tcf7−/− DN3 clones at a subset of CTCF+TCF-1 co-bound, as well as TCF-1-only peaks that gain insulation in Tcf7−/− DN3s. The two classes of peaks were defined using TCF-1 and CTCF ChIP-seq in thymocytes and DPs, respectively as in Fig. 1. These peaks were further selected based on the decrease of average interactions between the upstream and downstream of the peaks by at least 0.05 after TCF-1 deletion. The numbers are the average observed/expected interactions in the upper right square, which is the interactions between the upstream and downstream of the peaks. c. Local pileup interactions in wildtype and Bcl11b deficient naïve CD4 T+ cells at the CTCF and TCF-1 co-bound, as well as TCF-1 only peaks.
Extended Data Fig. 9 Oligopaint 3D FISH corroborates the role of TCF-1 on chromatin interactions at the Cd8 locus.
a-b. Contact matrix plot of Hi-C data sets at the Cd8a-Cd8b1 locus in NIH3T3 fibroblasts (a) and DN3s (b) (n=7221 genomic interactions). Inter-domain interactions were quantified by boxplots. Two sided Paired wilcoxon ranksum test was performed. Oligopaint probes for TAD1 (green) and TAD2 (magenta) are depicted. Two-sided Wilcoxon rank-sum test P-values are shown for boxplots. c-d. Box plot showing spatial distance (c) and spatial overlap (d) between TAD1 and TAD2 compared between wildtype (N=2) and TCF-1-deficient (N=3) of DPs in biological mouse replicates. Spatial distance and overlap were calculated as in (Fig. 5d, e). Number of cells used for imaging analysis per mouse is depicted in parenthesis. a-d Data are shown as boxplots (center, median; box limits, upper (75th) and lower (25th) percentiles; whiskers, 1.5× interquartile range; points, outliers).
Extended Data Fig. 10 Recruitment of cohesin at TCF-1 binding sites.
a. Volcano plot shows the differential CTCF binding in wildtype and Tcf7−/− DN3s. Red indicates stronger binding after TCF-1 deletion. Log2 fold change and P values are calculated with DEseq2. P depicts adjusted pvalue calculated by DESeq2. b. Volcano plot showing the differential SMC1 binding in wildtype and Tcf7−/− DN3s. Log2 fold change and P values were calculated with DEseq2. P depicts adjusted pvalue calculated by DESeq2. Red indicates stronger binding after TCF-1 deletion. c. Enriched motifs from HOMER at the gained or lost SMC1 peaks after TCF-1 deletion in DN3 cells using random background. The most significantly enriched motifs and associated P values are shown. P values were calculated using hypergenometric test. d. Cumulative distribution of genomic distance between 2,042 TCF-1 dependent SMC1 and NIPBL co-bound enhancers or 2,042 randomly selected genomic regions to 988 dynamic boundaries with TCF-1 and CTCF co-binding. Kolmogorov-Smirnov test P-value is shown for cumulative distribution plot.
Supplementary information
Supplementary Information
Supplementary Figs. 1–4
Supplementary Tables 1–16
Supplementary Table 1. TCF-1 and CTCF peak sets. TCF-1 and CTCF co-bind, TCF-1-alone and CTCF-alone sites in each sheet. Supplementary Table 2. T cell developmental genes from all stages downloaded from Kernfeld et al.28 Supplementary Table 3. Signature genes in each T cell development stage. T cell developmental genes in each stage downloaded from Kernfeld et al.28 Supplementary Table 4. Coordinates of different groups of boundaries. Boundaries from six developmental stages were grouped based on their presence in each stage. Supplementary Table 5. DEseq2 result of RNA-seq in untreated and TCF-1-induced fibroblasts. Differential genes are determined by log2(fold change) and P values are calculated from DEseq2. Supplementary Table 6. DEseq2 result of RNA-seq in WT and TCF-1 knockout (KO) DN3 cells. Differential genes are determined by log2(fold change) and P values are calculated from DEseq2. Supplementary Table 7. DEseq2 results of CTCF ChIP-seq in WT and TCF-1 KO DN3 cells. Differential peaks are determined by log2(fold change) and P values are calculated from DEseq2. Supplementary Table 8. DEseq2 result of SMC1 ChIP-seq in WT and TCF-1 KO DN3 cells. Differential peaks are determined by log2(fold change) and P values are calculated from DEseq2. Supplementary Table 9. Unique loops in WT and TCF-1 KO DN3 cells. Supplementary Table 10. Genes with RNA-seq log2(fold change) and Padj in the TADs that gain or lose intra-TAD interactions after TCF-1 KO. Log2(fold change) and Padj are calculated from DEseq2. Supplementary Table 11. Genes in the TADs that gain or lose intra-TAD interactions after TCF-1 KO. Supplementary Table 12. The subset of TCF-1 and CTCF co-bound sites or TCF-1-alone sites that lose insulation after TCF-1 overexpression in fibroblasts. Supplementary Table 13. The subset of TCF-1 and CTCF co-bound sites or TCF-1-alone sites that gain insulation after TCF-1 KO in DN3 cells. Supplementary Table 14. The subset of TCF-1 and CTCF co-bound sites or TCF-1-alone sites that gain insulation after TCF-1 KO in DP T cells. Supplementary Table 15. Genomic regions where TCF-1 led to a loss on the binding events of NIPBL and SMC1 proteins in addition to deposition of H3K27ac in DN3 T cells. Supplementary Table 16. Genomic regions where TCF-1 led to a gain on the binding events of NIPBL and SMC1 proteins in addition to deposition of H3K27ac in DN3 T cells
Source data
Source Data Extended Data Fig. 7
Unprocessed immunoblots related to Extended Data Fig. 7.
Rights and permissions
About this article
Cite this article
Wang, W., Chandra, A., Goldman, N. et al. TCF-1 promotes chromatin interactions across topologically associating domains in T cell progenitors. Nat Immunol 23, 1052–1062 (2022). https://doi.org/10.1038/s41590-022-01232-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-022-01232-z
This article is cited by
-
Dysregulation of Wnt/β-catenin signaling contributes to intestinal inflammation through regulation of group 3 innate lymphoid cells
Nature Communications (2024)
-
FOXO1 is a master regulator of memory programming in CAR T cells
Nature (2024)
-
Intrinsically disordered domain of transcription factor TCF-1 is required for T cell developmental fidelity
Nature Immunology (2023)
-
T cells in health and disease
Signal Transduction and Targeted Therapy (2023)
-
Three-dimensional genome organization in immune cell fate and function
Nature Reviews Immunology (2023)