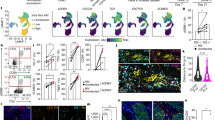
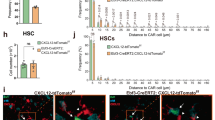
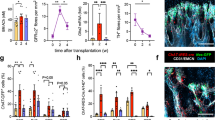
Abstract
Autonomic nerves control organ function through the sympathetic and parasympathetic branches, which have opposite effects. In the bone marrow, sympathetic (adrenergic) nerves promote hematopoiesis; however, how parasympathetic (cholinergic) signals modulate hematopoiesis is unclear. Here, we show that B lymphocytes are an important source of acetylcholine, a neurotransmitter of the parasympathetic nervous system, which reduced hematopoiesis. Single-cell RNA sequencing identified nine clusters of cells that expressed the cholinergic α7 nicotinic receptor (Chrna7) in the bone marrow stem cell niche, including endothelial and mesenchymal stromal cells (MSCs). Deletion of B cell-derived acetylcholine resulted in the differential expression of various genes, including Cxcl12 in leptin receptor+ (LepR+) stromal cells. Pharmacologic inhibition of acetylcholine esterase decreased the systemic supply of inflammatory myeloid cells in mice and humans with cardiovascular disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Change history
15 June 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41590-022-01266-3
References
Lahiri, M. K., Kannankeril, P. J. & Goldberger, J. J. Assessment of autonomic function in cardiovascular disease: physiological basis and prognostic implications. J. Am. Coll. Cardiol. 51, 1725–1733 (2008).
Mancia, G. & Grassi, G. The autonomic nervous system and hypertension. Circ. Res. 114, 1804–1814 (2014).
Florea, V. G. & Cohn, J. N. The autonomic nervous system and heart failure. Circ. Res. 114, 1815–1826 (2014).
Herring, N., Kalla, M. & Paterson, D. J. The autonomic nervous system and cardiac arrhythmias: current concepts and emerging therapies. Nat. Rev. Cardiol 16, 707–726 (2019).
Fujii, T. et al. Physiological functions of the cholinergic system in immune cells. J. Pharm. Sci. 134, 1–21 (2017).
Heeschen, C. et al. Nicotine stimulates angiogenesis and promotes tumor growth and atherosclerosis. Nat. Med. 7, 833–839 (2001).
Corradi, J. & Bouzat, C. Understanding the bases of function and modulation of α7 nicotinic receptors: implications for drug discovery. Mol. Pharmacol. 90, 288–299 (2016).
Wang, H. et al. Nicotinic acetylcholine receptor α7 subunit is an essential regulator of inflammation. Nature 421, 384–388 (2003).
Tracey, K. J. The inflammatory reflex. Nature 420, 853–859 (2002).
Katayama, Y. et al. Signals from the sympathetic nervous system regulate hematopoietic stem cell egress from bone marrow. Cell 124, 407–421 (2006).
Maryanovich, M., Takeishi, S. & Frenette, P. S. Neural regulation of bone and bone marrow. Cold Spring Harb. Perspect. Med 8, a031344 (2018).
Chang, R. B., Strochlic, D. E., Williams, E. K., Umans, B. D. & Liberles, S. D. Vagal sensory neuron subtypes that differentially control breathing. Cell 161, 622–633 (2015).
Artico, M. et al. Noradrenergic and cholinergic innervation of the bone marrow. Int. J. Mol. Med. 10, 77–80 (2002).
Santisteban, M. M., Zubcevic, J., Baekey, D. M. & Raizada, M. K. Dysfunctional brain–bone marrow communication: a paradigm shift in the pathophysiology of hypertension. Curr. Hypertens. Rep. 15, 377–389 (2013).
Jiang, W. et al. Acetylcholine-producing NK cells attenuate CNS inflammation via modulation of infiltrating monocytes/macrophages. Proc. Natl Acad. Sci. USA 114, E6202–E6211 (2017).
Reardon, C. et al. Lymphocyte-derived ACh regulates local innate but not adaptive immunity. Proc. Natl Acad. Sci. USA 110, 1410–1415 (2013).
Rosas-Ballina, M. et al. Acetylcholine-synthesizing T cells relay neural signals in a vagus nerve circuit. Science 334, 98–101 (2011).
Hirsch, N. P. Neuromuscular junction in health and disease. Br. J. Anaesth. 99, 132–138 (2007).
Kiel, M. J. et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121, 1109–1121 (2005).
Rosas-Ballina, M. et al. Splenic nerve is required for cholinergic antiinflammatory pathway control of TNF in endotoxemia. Proc. Natl Acad. Sci. USA 105, 11008–11013 (2008).
Costantini, T. W. et al. Uniquely human CHRFAM7A gene increases the hematopoietic stem cell reservoir in mice and amplifies their inflammatory response. Proc. Natl Acad. Sci. USA 116, 7932–7940 (2019).
Tikhonova, A. N. et al. The bone marrow microenvironment at single-cell resolution. Nature 569, 222–228 (2019).
Ding, L. & Morrison, S. J. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature 495, 231–235 (2013).
Tzeng, Y. S. et al. Loss of Cxcl12/Sdf-1 in adult mice decreases the quiescent state of hematopoietic stem/progenitor cells and alters the pattern of hematopoietic regeneration after myelosuppression. Blood 117, 429–439 (2011).
Arai, F. et al. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell 118, 149–161 (2004).
Ding, L., Saunders, T. L., Enikolopov, G. & Morrison, S. J. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 481, 457–462 (2012).
Dutta, P. et al. Myocardial infarction activates CCR2+ hematopoietic stem and progenitor cells. Cell Stem Cell 16, 477–487 (2015).
Beerman, I. et al. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc. Natl Acad. Sci. USA 107, 5465–5470 (2010).
Bernitz, J. M., Kim, H. S., MacArthur, B., Sieburg, H. & Moore, K. Hematopoietic stem cells count and remember self-renewal divisions. Cell 167, 1296–1309 (2016).
Dykstra, B., Olthof, S., Schreuder, J., Ritsema, M. & de Haan, G. Clonal analysis reveals multiple functional defects of aged murine hematopoietic stem cells. J. Exp. Med. 208, 2691–2703 (2011).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Soneson, C., Love, M. I. & Robinson, M. D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4, 1521 (2015).
Cunningham, F. et al. Ensembl 2019. Nucleic Acids Res. 47, D745–D751 (2019).
McCarthy, D. J., Chen, Y. & Smyth, G. K. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012).
Mootha, V. K. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Baryawno, N. et al. A cellular taxonomy of the bone marrow stroma in homeostasis and leukemia. Cell 177, 1915–1932 (2019).
Acknowledgements
We thank M. Handley, D. Daly, J. Kauffman, P. Sen, G. Lima, J. Choi and E. Surette of the HSCI-CRM Flow Cytometry Core Facility, Massachusetts General Hospital, for their assistance with cell sorting; the Bioanalytics Core at the Diabetes and Obesity Center, Christina Lee Brown Envirome Institute, University of Louisville, for mass spectrometry analysis; the BPF Next-Gen Sequencing Core Facility at Harvard Medical School for their expertise and instrument availability in support of this work and the Partners Healthcare Research Patient Data Registry group for facilitating the use of their database. We thank K. Joyes for editing the article. Figures were designed using Servier Medical Art (http://www.servier.com). This work was supported in part by the National Institutes of Health grants HL142494, NS108419, HL139598, HL125428, HL155097, HL149647, HL158040 and T32HL076136 and the MGH Research Scholar program. M.J.S. and D.R. were funded by Deutsche Forschungsgemeinschaft (SCHL 2221/1-1 and RO5071/1-1). M.H. was supported by an American Heart Association Career Development Award (19CDA34490005). C.S.M. was funded by NIH K99HL151750 and a Canadian Institutes of Health Research Banting Fellowship. J.M.H. and B.H.F. were supported by a grant from the One Brave Idea Initiative. G.S.M. was funded by the Fundação Lemann. The University of Louisville Diabetes and Obesity Center was supported by NIH P30 GM127607 and user fees. The Microscopy Core facility of the Massachusetts General Hospital Program in Membrane Biology receives support from Boston Area Diabetes and Endocrinology Research Center grant DK57521 and Center for the Study of Inflammatory Bowel Disease grant DK43351.
Author information
Authors and Affiliations
Contributions
M.J.S. and M.N. conceptualized, conceived and designed the study, interpreted data and made the figures. M.J.S., M.H., D.R., I.-H.L., N.S., B.H.F., S.Z., K.D.K., V.F., G.C., F.E.P., C.Y., Y.I., A.S.K., C.S.M., M.Y., S.P.S., G.R.W., G.S.M., K.G. and D.C. performed experiments and collected data. B.H.F., M.J.S., J.M.H. and M.N. analyzed and interpreted human data. M.J.S. performed mouse surgeries. M.J.S., M.H., I.-H.L., K.N. and M.N. designed, analyzed and processed RNA-sequencing data. M.J.S., M.H., D.R., I.-H.L., N.S., B.H.F., V.F., G.C., K.D.K., D.B., J.M.H., D.T.S., P.L., F.K.S., K.N. and M.N. discussed results and strategy. M.J.S. and M.N. wrote the manuscript with input from all authors. M.N. conceived and directed the study.
Corresponding author
Ethics declarations
Competing interests
M.N. has received funds or material research support from Alnylam, Biotronik, CSL Behring, GlycoMimetics, GSK, Medtronic, Novartis and Pfizer as well as consulting fees from Biogen, Gimv, IFM Therapeutics, Molecular Imaging, Sigilon and Verseau Therapeutics. The other authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks Paul Frenette and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Ioana Visan, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Acetylcholine and hematopoiesis.
a. 2,687 patients received either no donepezil (n = 870), 5 mg (n = 996) or 10 mg (n = 821) donepezil daily. Mean change in circulating white red blood cell counts (RBC) and platelets between 4 weeks before and 3 months after treatment. RBC: P = 0.0453 for no donepezil group vs. 10 mg donepezil group. Platelets: P = 0.0001 for no donepezil group vs. 10 mg donepezil group and P = 0.0001 for 5 mg donepezil group vs. 10 mg donepezil group. One-way ANOVA with Sidak’s post hoc test. Box shows 25th, 50th and 75th percentile but no whiskers or outliers. Blue dot represents the mean. b. Immunofluorescence image of a naive ChatGFP mouse femur. Section was stained with DAPI (blue; cell nucleus). Arrows indicate ChatGFP-positive nerves outside the bone marrow. The experiment was repeated two independent times with a similar result. Scale bar upper panel, 1 mm. All other panels, scale bar, 70 µm. Data are mean ± SEM with individual values for data distribution, *P < 0.05, and ****P < 0.0001.
Extended Data Fig. 2 Bone marrow acetylcholine is produced by mature B cells.
a. Flow cytometry for Chat-GFP expression in mature B cells and B cell progenitors in the bone marrow of ChatGFP mice. b. Flow cytometry gating for Chat-GFP expression by mature B cells in the bone marrow of ChatGFP mice and C57BL/6 mice. c. Flow cytometry plots of GFP+ B-2 cells in the bone marrow of ChatGFP mice. d-h. Flow cytometry plots of GFP expression by bone marrow cells in ChatGFP mice. i. Chat mRNA levels assessed in ChatGFP mice by qPCR in FACS-isolated mature GFP+ and GFP−B cells, CD45−/Ter119− niche cells, MSC, arteriolar endothelial cells (aECs) and sinusoidal EC (sEC). P = 0.0001 for ChatGFP+ B cell group vs. ChatGFP- B cell group. n = 11 mice per ChatGFP+ B cell group and n = 10 mice per ChatGFP- B cell group, two-tailed Student’s t-test. j. Intravital microscopy of the skull obtained in a ChatGFP mouse. Endothelial cells are labeled by injecting CD31/Sca-1 and B cells by B220. Arrows indicate GFP+ B220+ B cells. The experiment was repeated three independent times with a similar result. Scale bar, 10 µm. k. Confocal microscopy of the femur in ChatGFP mice (green). Endothelial cells are labeled by injecting CD31/ Sca-1 (blue). B220 control staining was performed using an isotype antibody (red). The experiment was performed once. Scale bar, 15 µm. l. Flow cytometry dot plots of cell cycle analysis of immature B cells in bone marrow of ChatGFP mice. m. Quantification of immature B cells in S/M/G2 cell cycle phase. P = 0.0040. n = 3 mice per group, two-tailed Student’s t-test. n. Flow cytometry plots of ChatGFP expression by circulating mature B cells in blood of ChatGFP mice. o. Percentage and numbers of GFP+ B cells in blood of ChatGFP mice. P = 0.0006 for percentage and P = 0.0023 for numbers of GFP+ B cells. Two independent experiments, n = 7 mice per group, two-tailed Mann−Whitney test. Data are mean ± SEM with individual values for data distribution, **P < 0.01, ***P < 0.001 and ****P < 0.0001.
Extended Data Fig. 3 Bone marrow acetylcholine preserves SLAM LSK stemness by reducing proliferation.
a. Bodyweight of Cd19CreChatfl/fl or their Chatfl/fl mice in steady-state. n = 8 mice per group, two-tailed Student’s t-test. Blood pressure in Cd19CreChatfl/fl or their Chatfl/fl mice in steady-state for 5 consecutive times and each individual measurement value is plotted. n = 8 mice per group, two-tailed Student’s t-test. Heart rate of Cd19CreChatfl/fl or Chatfl/fl mice. n = 8 mice per group, two-tailed Student’s t-test. b. Cell cycle of HPSCs was analyzed by flow cytomery in Cd19CreChatfl/fl or Chatfl/fl mice. Left, flow cytometry plots for cell cycle analysis of LSK and SLAM LSK. Right, cell cycle analysis of LSK and SLAM LSK by flow cytometry. SLAM LSK: P = 0.021 for G0-phase, P = 0.034 for G1-phase and P = 0.038 for S/G2/M-phase. LSK: P = 0.0002 for G0-phase, P = 0.0012 for G1-phase and P = 0.0003 for S/G2/M-phase. SLAM LSK: n = 8 mice per group, LSK: n = 7 Chatfl/fl, n = 8 Cd19CreChatfl/fl mice, two-tailed Student’s t-test. c. Limiting dilution assay. SLAM LSK from Cd19CreChatfl/fl or Chatfl/fl mice were FACS sorted and mixed with 3×105 BMNC from CD45.1 mice. Cell dilutions were transplanted into lethally irradiated CD45.1 recipients. Multilineage blood chimerism of 0.1% served as a cut-off to determine responders. d. Left table lists dilution steps, numbers of mice analyzed 4 months after transfer, and number of responders. Right table indicates HSC frequencies and 95% confidence interval (P = 0.452 for difference in frequency). e. Cd19CreChatfl/fl or Chatfl/fl mice were injected with Evans blue (20 mg/kg; i.v.) and tibias were harvested 4 hours thereafter. Vascular permeability by Evans blue quantification in the tibia. P = 0.0487. n = 7 Chatfl/fl, n = 6 Cd19CreChatfl/fl mice, two-tailed Student’s t-test. Data are mean ± SEM, n.s. P > 0.05, *P < 0.05, **P < 0.01 and ***P < 0.001.
Extended Data Fig. 4 B cell-derived acetylcholine effects in the spleen.
a. Membranes of the cytokine antibody array of the spleen, serum and bone marrow of naive Cd19CreChatfl/fl mice or their Chatfl/fl littermates. Membranes represent two independently performed experiments per group. b. Left, flow cytometry for splenic HSPC. Right, splenic HSPC enumeration in naive Cd19CreChatfl/fl or Chatfl/fl mice. P = 0.036 for SLAM LSK, P = 0.040 for LSK. n = 7 mice per group, two-tailed Student’s t-test. c. Leukocytes in spleen. P = 0.017 for monocytes, P = 0.038 for neutrophils. n = 7 mice per group, two-tailed Mann-Whitney test. d. Vascular permeability was analyzed by Evans blue extraction and quantification in the spleen of Cd19CreChatfl/fl mice or Chatfl/fl mice. P = 0.0455. n = 5 mice per group, two-tailed Student’s t-test. Data are mean ± SEM with individual values for data distribution, *P < 0.05.
Extended Data Fig. 5 Splenectomy and hematopoiesis.
a. Cd19CreChatfl/fl mice or their Chatfl/fl littermates underwent splenectomy or sham-operation and the bone marrow was harvested 4 weeks later for flow cytometry and cytokine/chemokine antibody array analysis. Membranes of the cytokine/chemokine antibody array of the bone marrow supernatant of splenectomized or sham-operated Cd19CreChatfl/fl mice or their Chatfl/fl littermates. Membranes represent two independently performed experiments per group. b. Flow cytometry plots for HSPC in the femur. c. Bone marrow HSPC enumeration in the femur. SLAM LSK: P = 0.0040 for sham, P = 0.0051 for splenectomy, GMP: P = 0.0003 for sham, P = 0.0108 for splenectomy, CMP: P = 0.0071 for sham, P = 0.0037 for splenectomy. Two independent experiments, n = 8 Chatfl/fl mice, sham Cd19CreChatfl/fl mice and splenectomy Chatfl/fl mice; n = 9 splenectomy Cd19CreChatfl/fl mice, two-tailed Student’s t-test. d. Flow cytometry plots for BrdU incorporation. e. Bone marrow HSPC proliferation analyzed by BrdU incorporation using flow cytometry. SLAM LSK: P = 0.0012 for sham, P = 0.0068 for splenectomy, GMP: P = 0.0034 for sham, P = 0.019 for splenectomy. Two independent experiments, n = 6 sham Chatfl/fl mice, n = 8 sham Cd19CreChatfl/fl mice, n = 7 splenectomy Chatfl/fl mice; n = 10 splenectomy Cd19CreChatfl/fl mice, two-tailed Student’s t-test. Data are mean ± SEM with individual values for data distribution, *P < 0.05, **P < 0.01 and ***P < 0.001.
Extended Data Fig. 6 Chrna7+ expression by bone marrow cells.
a. Acetylcholine receptor mRNA levels in bone marrow MSCs, ECs and osteolineage cells. Deposited data from reference22. n.d., not detectable. Data are mean ± SEM with individual values for data distribution. b. Flow cytometry plots for Chrna7 expression on GFP+ immature and mature B cells in the bone marrow of ChatGFP mice. c-f. Flow cytometry plots for Chrna7 expression on leukocytes in the bone marrow of C57BL/6 mice.
Extended Data Fig. 7 Chrna7+ bone marrow stromal cells.
a. Dot plot of signature gene expression in Chrna7+ bone marrow cell clusters assessed by single cell RNA sequencing. Chrna7+ bone marrow stromal cells were pooled post-hoc from Chatfl/fl and Cd19Cre Chatfl/fl steady-state mice (15,590 non-hematopoietic cells; n = 8 mice). Color indicates z-score of average gene expression (red, high; blue, low), circle size indicates percent of cells expressing the gene. b. Cd19CreChatfl/fl mice or their Chatfl/fl littermates were treated with AMD3100 (5 mg/kg; s.c.) or vehicle and blood was collected 4 h after treatment for colony forming unit (c.f.u.) and flow cytometry analysis. Circulating c.f.u. assay expressed as fold change over vehicle-treated mice of same genotype. P = 0.0047. n = 7 mice per Chatfl/fl group and n = 6 per Cd19CreChatfl/fl group, two-tailed Mann-Whitney test. c. Blood leukocytes expressed as fold change over vehicle-treated mice of same genotype. P = 0.0024. n = 7 mice per Chatfl/fl group and n = 6 per Cd19CreChatfl/fl group, two-tailed Mann-Whitney test. d. Bone marrow supernatant was harvested from naive Cd19CreChatfl/fl mice or their Chatfl/fl littermates and Tnfα protein was analyzed by ELISA. P = 0.0024. n = 7 mice per Chatfl/fl group and n = 6 per Cd19CreChatfl/fl group, two-tailed Mann-Whitney test. e. Left, flow cytometry plots for gating of CD31+ endothelial cells (ECs) and LepR+ MSCs in the bone marrow. Right, flow cytometry histogram plots of Chrna7 expression on ECs (left) and LepR+ MSCs (right). Numbers in the histogram represent the mean fluorescence intensity of 3 mice per group. Bar graphs represent the percentage of Chrna7+ cells among ECs or LepR+ MSCs. Gates were set using an Chrna7 isotype control antibody. P = 0.0076 for LepR+ MSCs. n = 3 mice per group, two-tailed Student’s t test. Data are mean ± SEM with individual values for data distribution, *P < 0.05 and **P < 0.01.
Extended Data Fig. 8 B cell-derived acetylcholine in mice with atherosclerosis.
a. Cd19CreChatfl/fl mice or their Chatfl/fl littermates were intravenously injected with an adeno-associated virus 2/8 encoding hPCSK9 and fed a high-fat diet (HFD) for 12 weeks. Serum cholesterol levels after 12 weeks of HDF. Two independent experiments, n = 6 mice per Chatfl/fl group and n = 8 per Cd19CreChatfl/fl group, two-tailed Mann-Whitney test. b. Aortic roots were harvested from ChatGFP mice with atherosclerosis for flow cytometry and histology analysis. GFP+ B cell abundance in the femur and aorta of ChatGFP mice assessed by flow cytometry. P = 0.0038 absolute number data and P = 0.0094 for relative frequency data. n = 4 mice per group, two-tailed Student’s t test. c. Immunofluorescence image of the aortic roots in ChatGFP mice (green). Sections were stained with B220 (red) and DAPI (blue). The experiment was performed with 4 independent mice, which all showed a similar result. Scale bar, 200 µm. d. Higher magnification images of areas indicated in e. Arrow heads indicate ChatGFP-positive nerve fiber, arrows indicate B220+ B cells. The experiment was performed with 4 independent mice, which all showed a similar result. Scale bar in panel 1, 10 µm, all other panels 20 µm. Data are mean ± SEM with individual values for data distribution, **P < 0.01.
Extended Data Fig. 9 Splenic hematopoiesis after MI is curbed by B-cell derived acetylcholine.
a. Cd19CreChatfl/fl and Chatfl/fl mice were subjected to myocardial infarction (MI) and harvested 6 days later. HSPC in the spleen 6d after MI measured by c.f.u. assay. P = 0.0325. n = 5 mice per group, two-tailed Student’s t-test. b. Flow cytometry plots for splenic HSPC 6d after MI. c. Splenic HSPC enumeration 6d after MI. P = 0.047 for SLAM LSK, P = 0.0140 for LSK, P = 0.020 for CMP and P = 0.011 for GMP. n = 5 mice per group, two-tailed Student’s t-test. d. Leukocytes in spleen 6d after MI. n = 5 mice per group, two-tailed Student’s t-test. e. Cd19CreChatfl/fl and Chatfl/fl mice underwent splenectomy. 3 weeks after surgery, mice were subjected to MI and harvested 3 days later for flow cytometry. C57BL/6 mice were used as controls in the no splenectomy group. Flow cytometry plots for HSPC in the bone marrow 3 d after MI in splenectomized mice. f. Bone marrow HSPC enumeration. Two independent experiments, SLAM LSK: P = 0.030 for no splenectomy, P = 0.014 for splenectomy. CMP: P = 0.0095 for no splenectomy, P = 0.048 for splenectomy. SLAM LSK: n = 6 no splenectomy Chatfl/fl mice, n = 5 no splenectomy Cd19CreChatfl/fl mice, n = 8 splenectomy Chatfl/fl mice, n = 7 splenectomy Cd19CreChatfl/fl mice. CMP: n = 7 no splenectomy Chatfl/fl mice, n = 6 no splenectomy Cd19CreChatfl/fl mice, n = 8 splenectomy Chatfl/fl mice, n = 7 splenectomy Cd19Cre Chatfl/fl mice, two-tailed Student’s t-test. g. Myeloid cells and neutrophils in circulation 3 d after MI. Myeloid cells: P = 0.0053 for no splenectomy, P = 0.0009 for splenectomy. Neutrophils: P = 0.011 for no splenectomy, P = 0.016 for splenectomy. Two independent experiments, n = 12 no splenectomy Chatfl/fl mice, n = 10 no splenectomy Cd19CreChatfl/fl mice, n = 8 splenectomy Chatfl/fl mice, n = 7 no splenectomy Cd19CreChatfl/fl mice, two-tailed Student’s t-test. h. Flow cytometry plots of myeloid cells in the infarct myocardium 3 d after MI in splenectomized mice. i. Myeloid cell and neutrophil numbers in the infarct myocardium 3 d after MI. Myeloid cells: P = 0.037 for no splenectomy, P = 0.027 for splenectomy. Neutrophils: P = 0.0006 for no splenectomy, P = 0.041 for splenectomy. Two independent experiments, n = 12 no splenectomy Chatfl/fl mice, n = 9 no splenectomy Cd19CreChatfl/fl mice, n = 8 splenectomy Chatfl/fl mice, n = 7 no splenectomy Cd19CreChatfl/fl mice, two-tailed Student’s t-test. Data are mean ± SEM with individual values for data distribution, *P < 0.05, **P < 0.01 and ***P < 0.001.
Extended Data Fig. 10 Acetylcholinesterase inhibitor contains hematopoiesis and inflammation after MI.
a. C57BL/6 mice receiving either saline or pyridostigmine for 4 weeks were subjected to myocardial infarction (MI) and assayed 3 days after MI. Flow cytometry plots for HSPC. b. Bone marrow HSPC, CMP, GMP and MDP enumeration using flow cytometry. P = 0.019 for SLAM LSK, P = 0.039 for CMP, P = 0.0059 for GMP and P = 0.0057 for MDP. Two independent experiments, n = 12 saline and n = 11 mice pyridostigmine, two-tailed Student’s t-test. c. C.f.u. assay of bone marrow mononuclear cells (BMNCs) for granulocytes and macrophages progenitors (GMs). P = 0.040. n = 5 saline, n = 4 mice pyridostigmine group, two-tailed Mann-Whitney test. d. Circulating HSPC measured by c.f.u. assay. P = 0.0159. n = 5 saline, n = 4 mice pyridostigmine, two-tailed Mann-Whitney test. e. Flow cytometry plots for circulating myeloid cells. f. Leukocyte subsets in circulation. P = 0.0015 for leukocytes, P = 0.026 for monocytes, P = 0.046 for Ly6high monocytes and P = 0.026 for neutrophils. Two independent experiments, n = 13 saline, n = 11 pyridostigmine mice, two-tailed Student’s t-test. g. Flow cytometry plots of myeloid cells in the infarct myocardium 3 days after MI. h. Myeloid cell numbers in the infarct myocardium 3 days after MI. P = 0.0093 for myeloid cells, P = 0.0056 for neutrophils, P = 0.0460 for monocytes and P = 0.0140 for macrophages. Two independent experiments, n = 13 saline, n = 11 pyridostigmine mice, two-tailed Student’s t-test. i. Myeloid cell numbers in the remote myocardium 3 days after MI. P = 0.049 for macrophages. Two independent experiments, n = 13 saline, n = 11 pyridostigmine mice, two-tailed Student’s t-test. i. C57BL/6 mice receiving either saline or pyridostigmine for 4 weeks were subjected to MI and harvested 7 days after MI. Immunohistochemical staining and quantification of CD11b+ myeloid cells, collagen I, α-smooth muscle actin+ myofibroblasts and CD31+ vessels in the infarct border zone 7d after MI. Bar graphs show percentage of positive staining per region of interest (ROI). Each data point represents a ROI and 4 ROI were analyzed per mouse. P = 0.023 for CD11b and P = 0.0041 for collagen I. Two independent experiments, n = 4 saline, n = 3 pyridostigmine mice, two-tailed Student’s t-test. k. Survival after MI. P = 0.697. Two independent experiments, n = 9 saline, n = 10 pyridostigmine mice, Mantel–Cox log-rank test. Data are mean ± SEM with individual values for data distribution, *P < 0.05 and **P < 0.01.
Supplementary information
Supplementary Video 1
Confocal microscopy z stack of cortical bone from a ChatGFP mouse shows GFP+ leukocytes and a nerve-like GFP+ structure in the cortical bone.
Supplementary Video 2
Confocal microscopy z stack of bone marrow in a ChatGFP mouse shows GFP+ leukocytes and no nerve-like GFP+ structures.
Rights and permissions
About this article
Cite this article
Schloss, M.J., Hulsmans, M., Rohde, D. et al. B lymphocyte-derived acetylcholine limits steady-state and emergency hematopoiesis. Nat Immunol 23, 605–618 (2022). https://doi.org/10.1038/s41590-022-01165-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-022-01165-7
This article is cited by
-
Cholinergic signaling via the α7 nicotinic acetylcholine receptor regulates the migration of monocyte-derived macrophages during acute inflammation
Journal of Neuroinflammation (2024)
-
The roles of bone remodeling in normal hematopoiesis and age-related hematological malignancies
Bone Research (2023)
-
B cell-derived acetylcholine curbs haematopoiesis
Nature Reviews Immunology (2022)
-
B cells regulate hematopoietic stem cells via cholinergic signaling
Nature Immunology (2022)