Abstract
Chronic inflammation triggers compensatory immunosuppression to stop inflammation and minimize tissue damage. Studies have demonstrated that endoplasmic reticulum (ER) stress augments the suppressive phenotypes of immune cells; however, the molecular mechanisms underpinning this process and how it links to the metabolic reprogramming of immunosuppressive macrophages remain elusive. In the present study, we report that the helper T cell 2 cytokine interleukin-4 and the tumor microenvironment increase the activity of a protein kinase RNA-like ER kinase (PERK)-signaling cascade in macrophages and promote immunosuppressive M2 activation and proliferation. Loss of PERK signaling impeded mitochondrial respiration and lipid oxidation critical for M2 macrophages. PERK activation mediated the upregulation of phosphoserine aminotransferase 1 (PSAT1) and serine biosynthesis via the downstream transcription factor ATF-4. Increased serine biosynthesis resulted in enhanced mitochondrial function and α-ketoglutarate production required for JMJD3-dependent epigenetic modification. Inhibition of PERK suppressed macrophage immunosuppressive activity and could enhance the efficacy of immune checkpoint programmed cell death protein 1 inhibition in melanoma. Our findings delineate a previously undescribed connection between PERK signaling and PSAT1-mediated serine metabolism critical for promoting immunosuppressive function in M2 macrophages.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
RNA-seq and ChIP-seq results are available in the Gene Expression Omnibus database under accession nos. GSE165836 and GSE183287, respectively. Other data available on request to corresponding author. Source data are provided with this paper.
References
Wynn, T. A., Chawla, A. & Pollard, J. W. Macrophage biology in development, homeostasis and disease. Nature 496, 445–455 (2013).
McNelis, J. C. & Olefsky, J. M. Macrophages, immunity, and metabolic disease. Immunity 41, 36–48 (2014).
Murray, P. J. Macrophage polarization. Annu. Rev. Physiol. 79, 541–566 (2017).
Etchegaray, J.-P. & Mostoslavsky, R. Interplay between metabolism and epigenetics: a nuclear adaptation to environmental changes. Mol. Cell 62, 695–711 (2016).
Lio, C. J. & Huang, S. C. Circles of life: linking metabolic and epigenetic cycles to immunity. Immunology 161, 165–174 (2020).
Buck, M. D., Sowell, R. T., Kaech, S. M. & Pearce, E. L. Metabolic instruction of immunity. Cell 169, 570–586 (2017).
Li, X. et al. Navigating metabolic pathways to enhance antitumour immunity and immunotherapy. Nat. Rev. Clin. Oncol. 16, 425–441 (2019).
Bettigole, S. E. & Glimcher, L. H. Endoplasmic reticulum stress in Immunity. Annu. Rev. Immunol. 33, 107–138 (2015).
Grootjans, J., Kaser, A., Kaufman, R. J. & Blumberg, R. S. The unfolded protein response in immunity and inflammation. Nat. Rev. Immunol. 16, 469–484 (2016).
Song, M. & Cubillos-Ruiz, J. R. Endoplasmic reticulumsstress responses in intratumoral immune cells: implications for cancer immunotherapy. Trends Immunol. 40, 128–141 (2019).
Osorio, F. et al. The unfolded-protein-response sensor IRE-1α regulates the function of CD8α+ dendritic cells. Nat. Immunol. 15, 248–257 (2014).
Cubillos-Ruiz, J. R. et al. ER stress sensor XBP1 controls anti-tumor immunity by disrupting dendritic cell homeostasis. Cell 161, 1527–1538 (2015).
Mohamed, E. et al. The unfolded protein response mediator PERK governs myeloid cell-driven immunosuppression in tumors through inhibition of STING signaling. Immunity 52, 668–682.e7 (2020).
Song, M. et al. IRE1α–XBP1 controls T cell function in ovarian cancer by regulating mitochondrial activity. Nature 562, 423–428 (2018).
Cao, Y. et al. ER stress-induced mediator C/EBP homologous protein thwarts effector T cell activity in tumors through T-bet repression. Nat. Commun. 10, 1280 (2019).
Walter, P. & Ron, D. The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011).
Shan, B. et al. The metabolic ER stress sensor IRE1α suppresses alternative activation of macrophages and impairs energy expenditure in obesity. Nat. Immunol. 18, 519–529 (2017).
Batista, A. et al. IRE1α regulates macrophage polarization, PD-L1 expression, and tumor survival. PLoS Biol. 18, e3000687 (2020).
Wek, R. C. & Cavener, D. R. Translational control and the unfolded protein response. Antioxid. Redox Sign 9, 2357–2372 (2007).
Rainbolt, T. K., Saunders, J. M. & Wiseman, R. L. Stress-responsive regulation of mitochondria through the ER unfolded protein response. Trends Endocrinol. Metab. 25, 528–537 (2014).
Wortel, I. M. N., Meer, L. T., van der, Kilberg, M. S. & van Leeuwen, F. N. Surviving stress: modulation of ATF4-mediated stress responses in normal and malignant cells. Trends Endocrinol. Metab. 28, 794–806 (2017).
Gundra, U. M. et al. Alternatively activated macrophages derived from monocytes and tissue macrophages are phenotypically and functionally distinct. Blood 123, e110–e122 (2014).
Lavin, Y. et al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell 169, 750–765.e17 (2017).
Huang, S. C.-C. et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat. Immunol. 15, 846–855 (2014).
Guthrie, L. N. et al. Attenuation of PKR-like ER kinase (PERK) signaling selectively controls endoplasmic reticulum stress-induced inflammation without compromising immunological responses*. J. Biol. Chem. 291, 15830–15840 (2016).
Yang, F. et al. ER-stress regulates macrophage polarization through pancreatic EIF-2alpha kinase. Cell Immunol. 336, 40–47 (2019).
Pakos‐Zebrucka, K. et al. The integrated stress response. EMBO Rep. 17, 1374–1395 (2016).
Costa-Mattioli, M. & Walter, P. The integrated stress response: from mechanism to disease. Science 368, eaat5314 (2020).
Vitale, I., Manic, G., Coussens, L. M., Kroemer, G. & Galluzzi, L. Macrophages and metabolism in the tumor microenvironment. Cell Metab. 30, 36–50 (2019).
Jha, A. K. et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42, 419–430 (2015).
Liu, P.-S. et al. α-Ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat. Immunol. 18, 985–994 (2017).
Lebeau, J. et al. The PERK arm of the unfolded protein response regulates mitochondrial morphology during acute endoplasmic reticulum. Stress Cell Rep. 22, 2827–2836 (2018).
Cogliati, S., Enriquez, J. A. & Scorrano, L. Mitochondrial cristae: where beauty meets functionality. Trends Biochem. Sci. 41, 261–273 (2016).
Bravo, R. et al. Increased ER-mitochondrial coupling promotes mitochondrial respiration and bioenergetics during early phases of ER stress. J. Cell Sci. 124, 2143–2152 (2011).
Haschemi, A. et al. The sedoheptulose kinase CARKL directs macrophage polarization through control of glucose metabolism. Cell Metab. 15, 813–826 (2012).
Ma, E. H. et al. Serine Is an essential metabolite for effector T cell expansion. Cell Metab. 25, 345–357 (2017).
Locasale, J. W. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat. Rev. Cancer 13, 572–583 (2013).
Ducker, G. S. & Rabinowitz, J. D. One-carbon metabolism in health and disease. Cell Metab. 25, 27–42 (2017).
Harding, H. P. et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108 (2000).
Harding, H. P. et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633 (2003).
Halaby, M. J. et al. GCN2 drives macrophage and MDSC function and immunosuppression in the tumor microenvironment. Sci. Immunol. 4, eaax8189 (2019).
Mullarky, E. et al. Identification of a small molecule inhibitor of 3-phosphoglycerate dehydrogenase to target serine biosynthesis in cancers. Proc. Natl Acad. Sci. USA 113, 1778–1783 (2016).
Pacold, M. E. et al. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat. Chem. Biol. 12, 452–458 (2016).
Kory, N. et al. SFXN1 is a mitochondrial serine transporter required for one-carbon metabolism. Science 362, eaat9528 (2018).
Gao, X. et al. Serine availability influences mitochondrial dynamics and function through lipid metabolism. Cell Rep. 22, 3507–3520 (2018).
Hwang, I.-Y. et al. Psat1-dependent fluctuations in α-ketoglutarate affect the timing of ESC differentiation. Cell Metab. 24, 494–501 (2016).
Harris, N. L. & Loke, P. Recent advances in type-2-cell-mediated immunity: insights from helminth infection. Immunity 47, 1024–1036 (2017).
Schäfer, M. & Werner, S. Cancer as an overhealing wound: an old hypothesis revisited. Nat. Rev. Mol. Cell Biol. 9, 628–638 (2008).
Eming, S. A., Wynn, T. A. & Martin, P. Inflammation and metabolism in tissue repair and regeneration. Science 356, 1026–1030 (2017).
DeNardo, D. G. et al. CD4+ T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell 16, 91–102 (2009).
Yang, X. et al. ATF4 regulates CD4+ T cell immune responses through metabolic reprogramming. CellReports 23, 1754–1766 (2018).
Ma, L. et al. Control of nutrient stress-induced metabolic reprogramming by PKCζ in tumorigenesis. Cell 152, 599–611 (2013).
Gao, S. et al. PSAT1 is regulated by ATF4 and enhances cell proliferation via the GSK3β/β-catenin/cyclin D1 signaling pathway in ER-negative breast cancer. J. Exp. Clin. Cancer Res. 36, 179 (2017).
Yang, C. et al. Glutamine‐utilizing transaminases are a metabolic vulnerability of TAZ/YAP‐activated cancer cells. EMBO Rep. 19, e43577 (2018).
Martínez-Reyes, I. & Chandel, N. S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 11, 102 (2020).
Chisolm, D. A. et al. CCCTC-binding factor translates Interleukin 2- and α-ketoglutarate-sensitive metabolic changes in T cells into context-dependent gene programs. Immunity 47, 251–267.e7 (2017).
Zhang, Q. et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature 525, 389–393 (2015).
Pan, W. et al. The DNA methylcytosine dioxygenase Tet2 sustains immunosuppressive function of tumor-infiltrating myeloid cells to promote melanoma progression. Immunity 47, 284–297.e5 (2017).
Finkelman, F. D. et al. Anti-cytokine antibodies as carrier proteins. Prolongation of in vivo effects of exogenous cytokines by injection of cytokine-anti-cytokine antibody complexes. J. Immunol. 151, 1235–1244 (1993).
Ivanisevic, J. et al. Toward ’omic scale metabolite profiling: a dual separation-mass spectrometry approach for coverage of lipid and central carbon metabolism. Anal. Chem. 85, 6876–6884 (2013).
Roci, I. et al. Metabolite profiling and stable isotope tracing in sorted subpopulations of mammalian cells. Anal. Chem. 88, 2707–2713 (2016).
Gallart-Ayala, H. et al. A global HILIC-MS approach to measure polar human cerebrospinal fluid metabolome: exploring gender-associated variation in a cohort of elderly cognitively healthy subjects. Analytica Chim. Acta 1037, 327–337 (2018).
Dunn, W. B. et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 6, 1060–1083 (2011).
Gagnebin, Y. et al. Metabolomic analysis of urine samples by UHPLC-QTOF-MS: Impact of normalization strategies. Analytica Chim. Acta 955, 27–35 (2017).
Tsugawa, H., Kanazawa, M., Ogiwara, A. & Arita, M. MRMPROBS suite for metabolomics using large-scale MRM assays. Bioinformatics 30, 2379–2380 (2014).
Conza, G. D. et al. Tumor-induced reshuffling of lipid composition on the endoplasmic reticulum membrane sustains macrophage survival and pro-tumorigenic activity. Nat. Immunol. 22, 1403–1415 (2021).
Acknowledgements
We thank S. Adoro and A. Huang for providing Eif2ak3fl/fl and OT-I mice, respectively. We thank A. Huang, S. Adoro and G. Dubyak for helpful discussions, and H. Fujioka from the Cleveland Center for Membrane and Structural Biology for expert technical assistance. All the individuals above are affiliated with Case Western Reserve University. L.N.R is supported by the Immunology T32 Training Program (no. AI089474). P.-C.H. is supported in part by a postdoctoral fellowship provided by Ministry of Science and Technology, Taiwan. C.-W.J.L. is funded by the National Institutes of Health’s National Cancer Institute K22 award (no. K22CA241290), and startup funds from the Department of Microbial Infection and Immunity and Pelotonia Institute of Immuno-oncology at Ohio State University. J.I. is supported by a UNIL interdisciplinary grant. P.-C.H. is funded by the European Research Council starting grant (no. 802773-MitoGuide), SNSF project grants (no. 31003A_182470), the Cancer Research Institute (CLIP investigator award and Lloyd J. Old STAR award) and University of Lausanne (UNIL) interdisciplinary grant. S.C.-C.H. is funded by the Cancer Research Institute CLIP Investigator Award, the VeloSano Pilot Award, Case Comprehensive Cancer Center American Cancer Society pilot grants (nos. IRG-91-022-19 and IRG-16-186-21), Case GI SPORE DRP grant (no. 5P50CA150964-08) and the Cleveland Digestive Research Core Center pilot grant (no. 1P30DK097948).
Author information
Authors and Affiliations
Contributions
L.N.R. and S.C-C.H. conceived the study. L.N.R., H.Z., H.-Y.C, H.G.-A., W.C. and Y.K. performed the experiments. L.N.R, H.Z., P.-S.L., J.I., C.-W.J.L., P.-C.H. and S.C.-C.H. analyzed the data. Y.W., H.G.-A, P.-C.H. and A.A.-L. performed the bioinformatics analysis. L.N.R. and S.C-C.H. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
P.-C.H. is a member of the scientific advisory for Elixiron Immunotherapeutics. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks Navdeep Chandel and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. L. A. Dempsey was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
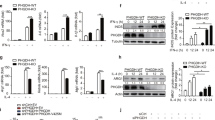
Extended Data Fig. 1 PERK deficiency does not affect M1 activation.
a, Enrichment plot of endoplasmic reticulum (ER) stress genes in IL-4c treated mouse peritoneal macrophages compared with naïve (PBS) macrophages by GSEA analysis. b, GSEA result comparing ER stress genes between TAMs and nontumor macrophages from patients with lung carcinoma. c, Pearson correlation of TAM CD68 expression with genes encoding molecules involved in the PERK pathway in caxncer patients from the TCGA. d, Immunoblot analysis of p-PERK, PERK, and β-actin from BMDMs stimulated with IL-4, Thapsigargin, or phosphatase. Ratio of p-PERK to total PERK was determined using ImageJ. Data are representative of three independent experiments. e, Frequency of p-PERK+ in BMDMs stimulated with IL-4 or Thapsigargin (n = 3; mean ± s.e.m). Data are collected from three independent experiments. f, Percentage of CD206+CD301+ in BMDMs treated with either DMSO (vehicle), IL-4, or Thapsigargin (n = 3; mean ± s.e.m). Data are collected from three independent experiments. g, Expression of CD206 and CD301 from BMDMs treated with IL-4 (M2) in the presence or absence of GSK2656157 (n = 3; mean ± s.e.m). Data are representative of three independent experiments. h, Percentage of 7AAD− naïve BMDMs isolated from Eif2ak3fl/fl or Eif2ak3fl/fl x LysMCre mice (n = 6; mean ± s.e.m). Data are collected from six independent experiments. i, Western blotting analysis of PERK, iNOS, Arg1, and β-actin from PERK wild-type and deficient BMDMs treated with LPS plus IFN-γ (M1), IL-4 (M2), or Thapsigargin. Data are representative of two independent experiments. j-l, Representative histogram (left) and quantitative plot (right) of iNOS (j), TNF-α (k), or expression of CD206 and CD301 (l) in BMDMs stimulated with LPS plus IFN-γ (M1) or IL-4 (M2) (n = 3; mean ± s.e.m). Data are representative of three independent experiments. m, Immunoblot analysis of p-PERK, PERK, XBP1s, and β-actin in unstimulated (M0), LPS + IFN-γ (M1), or IL-4 (M2) stimulated BMDMs. Data are representative of three independent experiments. All data were analyzed using two-tailed unpaired Student’s t-test (e,g,j,j,k,l) or one-way ANOVA with Tukey’s multiple comparisons test (f).
Extended Data Fig. 2 ISR in macrophage activation and metabolism.
a,b, Expression of CD206, CD301 (a), PD-L2 and Relmα (b) in macrophages stimulated with IL-4 in the presence or absence of ISRIB (n = 3; mean ± s.e.m). Data is representative of three independent experiments. c, Representative histogram (left) and quantitative analysis (right) of iNOS expression in macrophages stimulated with LPS + IFN-γ in the presence or absence of ISRIB (n = 3; mean ± s.e.m). Data is representative of three independent experiments. d, Expression of genes associated with ISR, assessed by RNA-Seq analysis. e,f, Expression of CD206, CD301 (e), PD-L2 and Relmα (f) in Gcn2+/+ and Gcn2-/- macrophages in the presence or absence of IL-4 (n = 3; mean ± s.e.m). Data is representative of two independent experiments. g,h, Representative histograms (left) and quantitative analysis (right) of iNOS (g) or TNF-α (h) in Gcn2+/+ and Gcn2-/- macrophages in the presence or absence of LPS + IFN-γ (n = 3; mean ± s.e.m). Data is representative of two independent experiments. i,j, Basal OCR (i) and ECAR (j) of Gcn2+/+ and Gcn2-/- M0, M1, and M2 macrophages (n = 3; mean ± s.e.m). Data is representative of two independent experiments. k, Basal OCR of M0 and M2 BMDMs in the presence or absence of ISRIB (n = 3; mean ± s.e.m). Data is representative of two independent experiments. l, Intracellular serine content from M0, M1 and M2 macrophages in the presence of absence of ISRIB (n = 3; mean ± s.e.m). Data is representative of two independent experiments. All data were analyzed using two-tailed unpaired Student’s t-test (a,b,c,e,f,g,h,I,j,k,l).
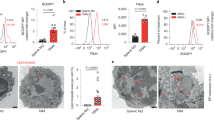
Extended Data Fig. 3 Pharmacological inhibition of PERK deviates cellular metabolism in M2 macrophages.
a, Basal OCR of naïve (M0) and M1 (LPS + IFN-γ) BMDMs from PERK sufficient or deficient mice (n = 3; mean ± s.e.m). Data collected from three independent experiments. b, Basal ECAR of naïve (M0), M1 (LPS + IFN-γ), and M2 (IL-4) BMDMs from PERK wild-type or null animals (n = 3; mean ± s.e.m). Data are collected from three independent experiments. c,d,e Basal OCR (c), ECAR (d), or ATP production (e) from wild-type BMDMs treated with IL-4 in the presence or absence of GSK2656157 (n = 3; mean ± s.e.m). Dashed line indicates wild-type M0. Data are collected from three independent experiments. f, Representative histogram (left) and quantitative analysis (right) of BODIPY FL C16 staining in BMDMs treated with IL-4 in the presence or absence of GSK2656157 (n = 3; mean ± s.e.m). Data representative of three independent experiments. g, Representative histogram (left) and the frequency (right) of BODIPY (493/503) staining in BMDMs stimulated with IL-4 in the presence or absence of GSK2656157 (n = 3; mean ± s.e.m). Data representative of three independent experiments. h, Representative TEM images of PERK wild-type and PERKcKO M2 (IL-4) macrophages. Red arrows indicate mitochondria. Data representative of three biological replicates. i, Representative histogram (left) and the frequency (right) of MitoTracker Green+ staining in BMDMs treated IL-4 (n = 3; mean ± s.e.m). Data representative of three independent experiments. j, Representative histogram (left) and the frequency (right) of MitoTracker Orange+ staining in BMDMs treated with IL-4 (n = 3; mean ± s.e.m). Data representative of three independent experiments. k, RNA-seq analysis of genes associated with mitochondrial calcium transport. l, Mitochondrial calcium flux (Rhod-2) from BMDMs treated with IL-4 in the presence or absence of GSK2656157 was determined and normalized by wild-type naïve M0 macrophages. Arrow, stimulation using 10 μM ionomycin (n = 6 for wild-type M0, M2; n = 4 for GSK-treated cells; mean ± s.e.m). Data are representative of three independent experiments. All data were analyzed using two-tailed unpaired Student’s t-test (a,b,c,d,e,f,g,i,j,l) or two-tailed paired Student’s t-test (l).
Extended Data Fig. 4 Inhibition of PHGDH or PSAT1 impairs M2 macrophage function.
a, Expression of genes encoding the serine one-carbon metabolic pathway in naïve (M0), M1 (LPS + IFN-γ), or M2 (IL-4) macrophages, assessed by RNA-Seq analysis. b,c, Western blotting analysis of PHGDH, PSAT1 and β-actin in BMDMs transduced with either retrovirus expressing Luc (b,c), PHGDH (b) or PSAT1 (c) shRNA and stimulated with IL-4. Data are representative of three independent experiments. d,e, Expression of CD206, CD301, PD-L2 and Relmα by IL-4 stimulated BMDMs treated with CBR-5884 (d) or transduced with PHGDH shRNA (e; middle), or transduced with PSAT1 shRNA (e; bottom) (n = 3; mean ± s.e.m). Data are representative of three independent experiments. f, Expression of CD206 and CD301 in M0 or LPS + IFN-γ (M1) stimulated BMDMs from Psat1fl/fl or Psat1fl/fl x LysMCre mice (n = 3; mean ± s.e.m). Data are representative of three independent experiments. g, Relative histogram (left) and gMFI (right) of either naïve (M0) or M1 macrophages from PSAT1 wild-type and knockout mice (n = 3; mean ± s.e.m). Data are representative of three independent experiments. h,i, Basal OCR and ECAR of wild-type BMDMs treated with IL-4 in the presence or absence of CBR-5884 (h) or transduced with shRNA targeting Phgdh or Psat1 (n = 3; mean ± s.e.m) (i). Data are collected from three independent experiments. j, ATP production from BMDMs stimulated with IL-4 in the presence or absence of CBR-5884 (n = 3; mean ± s.e.m). All data were analyzed using two-tailed unpaired Student’s t-test (d,f,g,h,j), or ordinary one-way ANOVA with Dunnett’s multiple comparisons test (e,i).
Extended Data Fig. 5 Loss of PERK does not result in H3K27m3 histone hypermethylation in pro-inflammatory M1 genes.
a,b, RT-qPCR analysis of Jmjd3 expression from PERK wild-type/knockout (a), or PSAT1 wild-type/knockout (b) BMDMs stimulated with IL-4 (n = 2; mean ± s.e.m). Data are collected from two independent experiments. c,d, Western blotting analysis of PERK, Jmjd3 and β-actin in M0, M1, and M2 BMDMs from PERK wild-type and knockout mice (c) or from PSAT1 wild-type and knockout mice (d). Data are representative two independent samples. e, Changes in gene expression of targets associated with H3K27me3 from PERK wild-type or knockout BMDMs stimulated with IL-4 (M2; left) or LPS + IFN-γ (M1; right). f, H3K27me3 histone modifications of Irf4, Pparg, Phgdh and Mgl2 from PERK wild-type and knockout BMDMs in the presence or absence of LPS + IFN-γ. g, H3K27me3 histone modifications of selected genes associated with pro-inflammation from naïve (M0), LPS + IFN-γ (M1) or IL-4 (M2) stimulated PERK wild-type or knockout macrophages. All data were analyzed using two-tailed unpaired Student’s t-test (a,b).
Extended Data Fig. 6 Therapeutic PERK inhibition in mouse melanoma model.
a, Schematic of the experimental design for small molecule inhibitor treatments. b, Body weight of tumor-bearing mice given the indicated treatments (n = 9 mice per group; mean ± s.e.m). Data are collected from two independent experiments. c, Frequency of tumor-infiltrating immune cells (TAMs, TILs, DCs, MDSCs, and NK cells) from B16-F10 tumor-bearing mice with treatment of either DMSO (Vehicle), GSK2656157, or NCT-503. Tumors were harvested on day 16. (n = 10 mice per group; mean ± s.e.m). Data are collected from two independent experiments. d, Schematic of the experimental design for the treatment of αPD-1, GSK2656157, and αPD-1 + GSK2656157. Data were analyzed by ordinary one-way ANOVA with Dunnett’s multiple comparisons test (b,c).
Supplementary information
Supplementary Information
Flow cytometry gating strategy.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 3
Unprocessed immunoblots.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Unprocessed immunoblots.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 6
Unprocessed immunoblots.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 7
Unprocessed immunoblots.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed immunoblots.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed immunoblots.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 5
Unprocessed immunoblots.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
About this article
Cite this article
Raines, L.N., Zhao, H., Wang, Y. et al. PERK is a critical metabolic hub for immunosuppressive function in macrophages. Nat Immunol 23, 431–445 (2022). https://doi.org/10.1038/s41590-022-01145-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-022-01145-x
This article is cited by
-
Metabolic regulation of tumor-associated macrophage heterogeneity: insights into the tumor microenvironment and immunotherapeutic opportunities
Biomarker Research (2024)
-
Emerging mechanisms of the unfolded protein response in therapeutic resistance: from chemotherapy to Immunotherapy
Cell Communication and Signaling (2024)
-
Targeting PHGDH reverses the immunosuppressive phenotype of tumor-associated macrophages through α-ketoglutarate and mTORC1 signaling
Cellular & Molecular Immunology (2024)
-
Serine metabolism in macrophage polarization
Inflammation Research (2024)
-
TOLLIP inhibits lipid accumulation and the integrated stress response in alveolar macrophages to control Mycobacterium tuberculosis infection
Nature Microbiology (2024)