Abstract
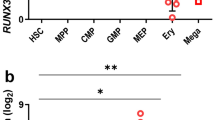
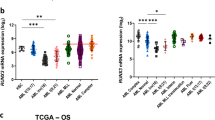
Anemia is a major comorbidity in aging, chronic kidney and inflammatory diseases, and hematologic malignancies. However, the transcriptomic networks governing hematopoietic differentiation in blood cell development remain incompletely defined. Here we report that the atypical kinase RIOK2 (right open reading frame kinase 2) is a master transcription factor (TF) that not only drives erythroid differentiation, but also simultaneously suppresses megakaryopoiesis and myelopoiesis in primary human stem and progenitor cells. Our study reveals the previously uncharacterized winged helix-turn-helix DNA-binding domain and two transactivation domains of RIOK2 that are critical to regulate key hematopoietic TFs GATA1, GATA2, SPI1, RUNX3 and KLF1. This establishes RIOK2 as an integral component of the transcriptional regulatory network governing human hematopoietic differentiation. Importantly, RIOK2 mRNA expression significantly correlates with these TFs and other hematopoietic genes in myelodysplastic syndromes, acute myeloid leukemia and chronic kidney disease. Further investigation of RIOK2-mediated transcriptional pathways should yield therapeutic approaches to correct defective hematopoiesis in hematologic disorders.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Raw data generated from proteomic analysis are deposited in MassIVE under accession code MSV000088238. Data generated from RNA-seq are deposited in the Gene Expression Omnibus (GEO) database under accession code GSE185922. ATAC–seq and ChIP–seq datasets are deposited in the GEO database under accession numbers GSE174714 and GSE174719, respectively. The mRNA correlations in individuals with MDS, AML and CKD have been drawn using publicly available GEO datasets under accession codes GSE19429, GSE131184 and GSE37171. This study did not generate new unique reagents. Requests for any materials (plasmids generated in this study, crRNAs) and reagents should be directed to and will be fulfilled by L.H.G. with appropriate material transfer agreements. Source data are provided with this paper.
References
Palapar, L. et al. Anaemia and physical and mental health in the very old: an individual participant data meta-analysis of four longitudinal studies of ageing. Age Ageing 50, 113–119 (2021).
Lopes, M. B. et al. A real-world longitudinal study of anemia management in non-dialysis-dependent chronic kidney disease patients: a multinational analysis of CKDopps. Sci. Rep. 11, 1784 (2021).
Becktell, K. et al. Aplastic Anemia & MDS International Foundation: Bone Marrow Failure Disease Scientific Symposium 2018. Leuk. Res. 80, 19–25 (2019).
Saygin, C. & Carraway, H. E. Current and emerging strategies for management of myelodysplastic syndromes. Blood Rev. 48, 100791 (2020).
Hong, S. et al. Survival following relapse after allogeneic hematopoietic cell transplantation for acute leukemia and myelodysplastic syndromes in the contemporary era. Hematol. Oncol. Stem Cell Ther. S1658-3876, 30178–3 (2020).
Garcia-Manero, G., Chien, K. S. & Montalban-Bravo, G. Myelodysplastic syndromes: 2021 update on diagnosis, risk stratification and management. Am. J. Hematol. 95, 1399–1420 (2020).
Feld, J., Navada, S. C. & Silverman, L. R. Myelo-deception: luspatercept & TGF-beta ligand traps in myeloid diseases & anemia. Leuk. Res. 97, 106430 (2020).
Bhatt, V. R. & Steensma, D. P. Hematopoietic cell transplantation for myelodysplastic syndromes. J. Oncol. Pract. 12, 786–792 (2016).
Ferreira-Cerca, S. et al. ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit. Nat. Struct. Mol. Biol. 19, 1316–1323 (2012).
Zemp, I. et al. Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J. Cell Biol. 185, 1167–1180 (2009).
Ameismeier, M., Cheng, J., Berninghausen, O. & Beckmann, R. Visualizing late states of human 40S ribosomal subunit maturation. Nature 558, 249–253 (2018).
Raundhal, M. et al. Blockade of IL-22 signaling reverses erythroid dysfunction in stress-induced anemias. Nat. Immunol. 22, 520–529 (2021).
Pellagatti, A. & Boultwood, J. The molecular pathogenesis of the myelodysplastic syndromes. Eur. J. Haematol. 95, 3–15 (2015).
Pellagatti, A. et al. Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells. Leukemia 24, 756–764 (2010).
Brennan, R. G. The winged-helix DNA-binding motif: another helix-turn-helix takeoff. Cell 74, 773–776 (1993).
LaRonde-LeBlanc, N. & Wlodawer, A. A family portrait of the RIO kinases. J. Biol. Chem. 280, 37297–37300 (2005).
Teichmann, M., Dumay-Odelot, H. & Fribourg, S. Structural and functional aspects of winged-helix domains at the core of transcription initiation complexes. Transcription 3, 2–7 (2012).
Nakahata, T. & Okumura, N. Cell surface antigen expression in human erythroid progenitors: erythroid and megakaryocytic markers. Leuk. Lymphoma 13, 401–409 (1994).
Tothova, Z. et al. Multiplex CRISPR–Cas9-based genome editing in human hematopoietic stem cells models clonal hematopoiesis and myeloid neoplasia. Cell Stem Cell 21, 547–555 (2017).
Khajuria, R. K. et al. Ribosome levels selectively regulate translation and lineage commitment in human hematopoiesis. Cell 173, 90–103 (2018).
Gutierrez, L., Caballero, N., Fernandez-Calleja, L., Karkoulia, E. & Strouboulis, J. Regulation of GATA1 levels in erythropoiesis. IUBMB Life 72, 89–105 (2020).
Gutierrez, L. et al. Ablation of Gata1 in adult mice results in aplastic crisis, revealing its essential role in steady-state and stress erythropoiesis. Blood 111, 4375–4385 (2008).
Fujiwara, T. GATA transcription factors: basic principles and related human disorders. Tohoku J. Exp. Med. 242, 83–91 (2017).
Ling, T. & Crispino, J. D. GATA1 mutations in red cell disorders. IUBMB Life 72, 106–118 (2020).
Gnanapragasam, M. N. & Bieker, J. J. Orchestration of late events in erythropoiesis by KLF1/EKLF. Curr. Opin. Hematol. 24, 183–190 (2017).
Zhang, D. E. et al. Function of PU.1 (Spi-1), C/EBP and AML1 in early myelopoiesis: regulation of multiple myeloid CSF receptor promoters. Curr. Top. Microbiol. Immunol. 211, 137–147 (1996).
Yokomizo-Nakano, T. et al. Overexpression of RUNX3 represses RUNX1 to drive transformation of myelodysplastic syndrome. Cancer Res. 80, 2523–2536 (2020).
Daw, S. & Law, S. The functional interplay of transcription factors and cell adhesion molecules in experimental myelodysplasia including hematopoietic stem progenitor compartment. Mol. Cell. Biochem. 476, 535–551 (2020).
Nerlov, C. & Graf, T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 12, 2403–2412 (1998).
Rekhtman, N. et al. PU.1 and pRB interact and cooperate to repress GATA-1 and block erythroid differentiation. Mol. Cell. Biol. 23, 7460–7474 (2003).
Shimizu, R. & Yamamoto, M. Quantitative and qualitative impairments in GATA2 and myeloid neoplasms. IUBMB Life 72, 142–150 (2020).
Huang, Z. et al. GATA-2 reinforces megakaryocyte development in the absence of GATA-1. Mol. Cell. Biol. 29, 5168–5180 (2009).
Aravind, L., Anantharaman, V., Balaji, S., Babu, M. M. & Iyer, L. M. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 29, 231–262 (2005).
Zhong, X. et al. HoxA9 transforms murine myeloid cells by a feedback loop driving expression of key oncogenes and cell cycle control genes. Blood Adv. 2, 3137–3148 (2018).
Xu, J. et al. Transcriptional silencing of γ-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev. 24, 783–798 (2010).
Chen, T. W. et al. ChIPseek, a web-based analysis tool for ChIP data. BMC Genomics 15, 539 (2014).
Harish, B., Swapna, G. V., Kornhaber, G. J., Montelione, G. T. & Carey, J. Multiple helical conformations of the helix-turn-helix region revealed by NOE-restrained MD simulations of tryptophan aporepressor, TrpR. Proteins 85, 731–740 (2017).
Bi, X. et al. RNA targets ribogenesis factor WDR43 to chromatin for transcription and pluripotency control. Mol. Cell 75, 102–116 (2019).
Chaparro, C. M. & Suchdev, P. S. Anemia epidemiology, pathophysiology, and etiology in low- and middle-income countries. Ann. N. Y. Acad. Sci. 1450, 15–31 (2019).
Feld, J., Belasen, A. & Navada, S. C. Myelodysplastic syndromes: a review of therapeutic progress over the past 10 years. Expert Rev. Anticancer Ther. 20, 465–482 (2020).
Groopman, J. E. & Itri, L. M. Chemotherapy-induced anemia in adults: incidence and treatment. J. Natl Cancer Inst. 91, 1616–1634 (1999).
Edginton-White, B. & Bonifer, C. The transcriptional regulation of normal and malignant blood cell development. FEBS J https://doi.org/10.1111/febs.15735 (2021).
Suzuki, M. et al. GATA factor switching from GATA2 to GATA1 contributes to erythroid differentiation. Genes Cells 18, 921–933 (2013).
Moriguchi, T. & Yamamoto, M. A regulatory network governing Gata1 and Gata2 gene transcription orchestrates erythroid lineage differentiation. Int. J. Hematol. 100, 417–424 (2014).
Szabo, S. J. et al. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100, 655–669 (2000).
Li, J. et al. Isolation and transcriptome analyses of human erythroid progenitors: BFU-E and CFU-E. Blood 124, 3636–3645 (2014).
Kaufman, D. S., Hanson, E. T., Lewis, R. L., Auerbach, R. & Thomson, J. A. Hematopoietic colony-forming cells derived from human embryonic stem cells. Proc. Natl Acad. Sci. USA 98, 10716–10721 (2001).
Myers, S. A. et al. Streamlined protocol for deep proteomic profiling of FAC-sorted cells and its application to freshly isolated murine immune cells. Mol. Cell Proteomics 18, 995–1009 (2019).
Wang, J., Varin, T., Vieth, M. & Elkins, J. M. Crystal structure of human RIOK2 bound to a specific inhibitor. Open Biol. 9, 190037 (2019).
Chen, C. S., White, A., Love, J., Murphy, J. R. & Ringe, D. Methyl groups of thymine bases are important for nucleic acid recognition by DtxR. Biochemistry 39, 10397–10407 (2000).
Acknowledgements
We thank V. Sankaran and R. Voit (Boston Children’s hospital) for their valuable input in primary human HSPC culture techniques and for providing HMD-empty vector and HMD-GATA1 expression plasmids; S. Badrinath, N. Mathewson and K. Wucherpfennig for their kind assistance in CRISPR–Cas9 genome editing and for providing EF1α-MCS-IRES-ZsGreen lentiviral expression vector; and M. Katsuyama (Kyoto Prefectural University of Medicine) for generously providing GATA2 and GATA3 overexpression plasmids. We also thank A. Wight for assistance in analyzing RNA-seq results. This work was funed by a Discovery Research Grant, the Edward P. Evans Foundation (to L.H.G.), the National Institutes of Health (NIH) small grant program (1R03HL156574-01 to L.H.G.), institutional funding, the Dana-Farber Cancer Institute (to L.H.G.), The Pussycat Foundation Helen Gurley Brown Presidential Initiative (to S.G.), National Cancer Institute (NCI) Clinical Proteomic Tumor Analysis Consortium grants (NIH/NCI U24-CA210986 and U01 CA214125 to S.A.C.) and NIH grants (R01HL146642 and R37CA228304 to X.C.).
Author information
Authors and Affiliations
Contributions
Conceptualization, S.G. and L.H.G.; design, S.G., G.A.P. and L.H.G.; methodology, S.G., S.A.M., S.A.C., X.C. and G.A.P.; data analysis, S.G., M.R., G.A.P. and L.H.G.; funding acquisition, S.G., M.R. and L.H.G.; writing–original draft, S.G., G.A.P. and L.H.G.; supervision, G.A.P. and L.H.G.
Corresponding authors
Ethics declarations
Competing interests
L.H.G. is a former Director of Bristol-Myers Squibb and the Waters Corporation and currently serves on the Board of Directors of GlaxoSmithKline Pharmaceuticals and Analog Devices and on the scientific advisory boards of Repare Therapeutics, Abpro Therapeutics and Kaleido Therapeutics. G.A.P. is on the Scientific Advisory Boards of Amicus Therapeutics, MeiraGTx, Annovis Bio, Retromer Therapeutics and Proclara Biosciences and holds equity or stock options in Denali Therapeutics, MeiraGTx, Annovis Bio, Retromer Therapeutics and Proclara Biosciences, companies that are developing therapies for neurodegenerative diseases. M.R. is currently an employee at Jnana Therapeutics. The other authors declare no competing interests.
Additional information
Peer review information Nature Immunology thanks Kazuhiko Igarashi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. L. A. Dempsey was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 RIOK2 drives erythropoiesis and concomitantly suppresses megakaryopoiesis and myelopoiesis.
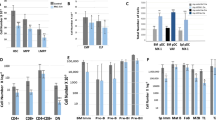
(a,b) Frequency of CD71 + CD235a- and CD71 + CD235a + population in control vs RIOK2 depleted TF-1 and K562 cells, respectively. (c,d) FACS plots and frequency of megakaryocytes (CD41/CD61 + ) after RIOK2 knockdown in TF-1 and K562 cells, respectively; sh#1 and 2: shRIOK2 #1 and #2. (e) Histogram plots depicting erythroblasts (CD235a), megakaryocytes (CD41/61) and myeloblasts (CD11b) in differentiating HSPCs after KD and KO of RIOK2. (f) Quantification of data presented in e, n = 4 independent donors. n = 3 technical replicates in a-d. ** p < 0.01, *** p < 0.001, **** p < 0.0001, one-way ANOVA with Tukey’s or Dunnett’s correction in c,d,f; unpaired two-tailed Student’s t-test in a,b. Data represented as mean ± s.e.m.
Extended Data Fig. 2 RIOK2 drives erythropoiesis and inhibits megakaryopoiesis and myelopoiesis.
(a,b) Total no. of myeloid (CD11b + ) and megakaryocytic (CD41/61 + ) cells after selective differentiation of control (Ctrl) vs RIOK2 KD and KO HSPCs to myeloid and megakaryocytic lineages, respectively. (c,d) Myeloid (CD13 + CD14 + ) and megakaryocytic (CD41 + CD42b + ) cells after selective differentiation of control (Ctrl) vs RIOK2 KD and KO HSPCs to myeloid and megakaryocytic lineages, respectively. (e) Images of blast forming unit-erythroid (BFU-E) and colony forming unit-granulocyte macrophage progenitors (CFU-GM) after KD and KO of RIOK2 in HSPCs, scale bar 750 µm. (f) Quantification of BFU-E, CFU-E and CFU-GM colonies in control vs RIOK2 KD and KO HSPCs. (g) Principal component analysis (PCA) of quantitative proteomic dataset showing control vs RIOK2 KD and KO primary human HSPCs, n = 3 donors. (h) Volcano plot showing differentially expressed proteins in control vs RIOK2 KD HSPCs. (i) Gene set enrichment analysis (GSEA) plot showing defective ribosome biogenesis in RIOK2 KO vs control HSPCs. n = 4 independent donors in a-d, f. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, one-way ANOVA with Tukey’s or Dunnett’s correction. Data represented as mean ± s.e.m. Fig. e representative of 4 independent experiments.
Extended Data Fig. 3 RIOK2 and GATA1 form a positive feedback loop to regulate hematopoietic differentiation.
(a) GATA binding motif (yellow) in the promoter region of RIOK2 (black) followed by the first exon (blue) and ATG start codon (red). (b) RIOK2 promoter-driven luciferase activity in response to increasing GATA1, GATA2 and GATA3 expression in HEK293 cells. (c) mRNA levels of RIOK2 (normalized to actin) after GATA1 overexpression (OE) and adjoining co-relation plot; EV: empty vector; Pearson’s correlation coefficient (r) and P value are shown. (d) RIOK2 mRNA expression (normalized to actin) in RIOK2 KD vs control cells. (e) GATA1 mRNA expression (normalized to actin) in GATA1 KD vs control cells. (f) HBB, ITGAM and ITGB3 mRNA expression (normalized to actin) in RIOK2 KD vs GATA1 KD HSPCs, compared to control. (g) Schema showing positive feedback loop between GATA1 and RIOK2 regulating hematopoietic differentiation. n = 3 independent donors in d-f; n = 4 and 6 technical replicates in b and c respectively. * p < 0.05, ** p < 0.01, *** p < 0.001, one-way ANOVA with Tukey’s correction in f, Unpaired two-tailed Student’s t-test in fig. c-e. Data represented as mean ± s.e.m.
Extended Data Fig. 4 RIOK2 regulates expression of transcription factors involved in hematopoietic lineage commitment.
(a) Volcano plots showing differentially expressed genes (cut off: adjusted P-Value<0.05) in control vs RIOK2 KD and control vs RIOK2 KO HSPCs; red: upregulated genes, blue: downregulated genes. (b) GATA2 mRNA level (normalized to actin) in control vs RIOK2 KD cells after suppression of GATA2 with 2 different crRNAs. (c) Erythroid progression (CD235a) in scrambled vs RIOK2 KD cells with or without suppression of GATA2 with 2 different crRNAs. (d) Megakaryocytic progression (CD41/61+ cells) in scrambled vs RIOK2 KD cells with or without knockdown of SPI1/RUNX3/GATA2 using 2 crRNAs against each. (e) Graph showing comparable mapped reads in control and RIOK2 KO HSPCs in ATAC-sequencing. n = 4 technical replicates in b-d. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, one-way ANOVA with Tukey’s or Dunnett’s correction. Data represented as mean ± s.e.m.
Extended Data Fig. 5 RIOK2 binds to a specific de novo nucleotide motif in the human genome.
(a) Relative binding intensity of RIOK2 at the transcription start site (TSS) in RIOK2 immunoprecipitated (RIOK2 IP) vs input sample. (b) Chromosome view plots depicting binding of RIOK2 at the promoters of GATA2 and SPI1. (c) De novo DNA binding sequences identified in the entire genome via ChIP sequencing using monoclonal antibodies of RIOK2. The sequence highlighted in red was identified in the promoters of RIOK2’s target genes. (d) Presence of de novo nucleotide binding motif specific for RIOK2 in the promoter regions of its putative target genes: GATA1, GATA2, RUNX3, SPI1, and KLF1.
Extended Data Fig. 6 RIOK2 binds DNA in vitro.
(a) Electrophoretic mobility shift assay (EMSA) showing WT (central CCC) or mutant (MUT: central CCC mutated to TTT) DNA migration in the presence of recombinant human RIOK2. (b) Quantitative band shift represented in a. (c,d) EMSA and adjoining quantification showing increasing DNA binding ability of RIOK2 over a time course; mins: minutes. (e,f) EMSA and adjoining quantification showing dose-dependent increase in DNA binding ability of RIOK2 with increasing protein concentration. (g) mRNA levels of early erythroid genes (HBB, HBA1, FECH, TFRC, KEL: normalized to actin) in RIOK2-KO HSPCs ectopically expressing EV, WT, DBM or NTE RIOK2, SCR: scrambled. n = 3 and 4 per group in d and f respectively. n = 3 experimental replicates in b,g. ** p < 0.01, **** p < 0.0001, ns: not significant, Unpaired two-tailed Student’s t-test in b, one-way ANOVA with Dunnett’s correction in g. Data represented as mean ± s.e.m. Fig. a,c representative of 3 independent experiments and fig. e representative of 4 independent experiments.
Extended Data Fig. 7 Characterization of the transactivation (TAD) and transrepressor (TRD) domains of RIOK2.
(a) ChIP of the promoter regions of GATA1, GATA2, RUNX3 and KLF1 by empty vector (EV) or HA-tagged wild-type (WT), ΔTAD1 (transactivation domain 1 deleted) and ΔTAD2 (transactivation domain 2 deleted) RIOK2 using anti-HA antibodies. (b) mRNA levels of early erythroid genes (HBB, HBA1, FECH, TFRC, KEL: normalized to actin) in RIOK2-KO HSPCs ectopically expressing EV, WT, ΔTAD1 or ΔTAD2 RIOK2, SCR: scrambled. (c) Erythroid progression (CD235a) in RIOK2-KO HSPCs ectopically expressing EV, WT or ΔTRD (deletion of transrepressor domain) RIOK2. (d) mRNA levels of early erythroid genes (HBA1, HBA2, SPTA1, TFRC: normalized to actin) in RIOK2-KO HSPCs ectopically expressing EV, WT or ΔTRD RIOK2. All comparisons done with respect to KO + EV group in fig. b-d. (e) Schema showing the 3 known domains of human RIOK2: N-terminal wHTH domain, central RIO domain and C-terminal domain. Positions of the DNA binding domain (DBD) and the transactivation domains (TAD1 and TAD2) of RIOK2 are shown. (f) Diagram illustrating RIOK2 as a master transcriptional regulator of key transcription factors (GATA1, KLF1, RUNX3, SPI1 and GATA2) in hematopoiesis. n = 2 per group in a; n = 3 independent donors in b-d. Data represented as mean ± SEM. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, ns: not significant, one-way ANOVA with Tukey’s or Dunnett’s correction in b-d. Data represented as mean ± s.e.m.
Extended Data Fig. 8 The kinase domain of RIOK2 is involved in erythropoiesis but does not affect RIOK2’s transcriptional activities.
(a) GATA1, SPI1 and RUNX3 promoter-driven luciferase reporter activity after expression of EV, WT, K123A, DBM or ΔTAD1 RIOK2; all comparisons done with respect to EV group (b) EMSA showing DNA-binding affinities of EV, WT, K123A, DBM, ΔTAD1 and ΔTAD2 RIOK2. (c) Western blot showing expression of ectopically expressed HA-tagged WT, DBM, K123A, ΔTAD1 RIOK2 or EV in RIOK2 KO HSPCs. (d) O-propargyl-puromycin (OPP) incorporation in control (Ctrl) vs RIOK2 KO cells after reconstitution of WT, DBM, K123A RIOK2 or EV. (e-g) Selective progression of erythroid (CD235a), myeloid (CD11b), and megakaryocytic (CD41/61) differentiation in HSPCs ectopically expressing EV, WT, DBM, ΔTAD1 or K123A RIOK2 in RIOK2-KO setting, respectively. All comparisons done with respect to EV in a, and KO + EV group in d-g. n = 3 technical replicates in a, d-g. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, ns: not significant, one-way ANOVA with Tukey’s or Dunnett’s correction. Data represented as mean ± s.e.m. Fig. b and c representative of 2 independent experiments.
Extended Data Fig. 9 Graphical illustration of RIOK2 functioning as a master transcription factor governing hematopoietic differentiation.
RIOK2 transcriptionally regulates key TFs in hematopoietic differentiation.
Extended Data Fig. 10 Flow cytometry plots showing gating strategies.
(a) erythroid, myeloid and megakaryocytic lineages (b) BFU-E and CFU-E formation (c) CFU-GM and CFU-Mk formation.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1
Unprocessed immunoblots.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 5
Unprocessed immunoblots.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 6
Unprocessed immunoblots.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 7
Unprocessed immunoblots
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 6
Unprocessed immunoblots.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 8
Unprocessed immunoblots and gels.
Rights and permissions
About this article
Cite this article
Ghosh, S., Raundhal, M., Myers, S.A. et al. Identification of RIOK2 as a master regulator of human blood cell development. Nat Immunol 23, 109–121 (2022). https://doi.org/10.1038/s41590-021-01079-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-021-01079-w
This article is cited by
-
Saracatinib prompts hemin-induced K562 erythroid differentiation but suppresses erythropoiesis of hematopoietic stem cells
Human Cell (2024)
-
Identification of a genomic DNA sequence that quantitatively modulates KLF1 transcription factor expression in differentiating human hematopoietic cells
Scientific Reports (2023)