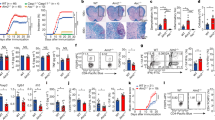
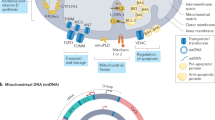
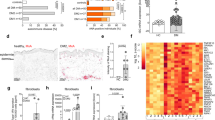
Abstract
Misdirected immunity gives rise to the autoimmune tissue inflammation of rheumatoid arthritis, in which excess production of the cytokine tumor necrosis factor (TNF) is a central pathogenic event. Mechanisms underlying the breakdown of self-tolerance are unclear, but T cells in the arthritic joint have a distinctive metabolic signature of ATPlo acetyl-CoAhi proinflammatory effector cells. Here we show that a deficiency in the production of mitochondrial aspartate is an important abnormality in these autoimmune T cells. Shortage of mitochondrial aspartate disrupted the regeneration of the metabolic cofactor nicotinamide adenine dinucleotide, causing ADP deribosylation of the endoplasmic reticulum (ER) sensor GRP78/BiP. As a result, ribosome-rich ER membranes expanded, promoting co-translational translocation and enhanced biogenesis of transmembrane TNF. ERrich T cells were the predominant TNF producers in the arthritic joint. Transfer of intact mitochondria into T cells, as well as supplementation of exogenous aspartate, rescued the mitochondria-instructed expansion of ER membranes and suppressed TNF release and rheumatoid tissue inflammation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All data in the present study are available within the article and its Supplementary files and from the corresponding author upon reasonable request. Source data are provided with this paper.
References
Deane, K. D., Norris, J. M. & Holers, V. M. Preclinical rheumatoid arthritis: identification, evaluation, and future directions for investigation. Rheum. Dis. Clin. North Am. 36, 213–241 (2010).
Feldmann, M., Brennan, F. M. & Maini, R. N. Role of cytokines in rheumatoid arthritis. Annu. Rev. Immunol. 14, 397–440 (1996).
Maini, R. N. & Taylor, P. C. Anti-cytokine therapy for rheumatoid arthritis. Annu. Rev. Med 51, 207–229 (2000).
Zhang, F. et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 20, 928–942 (2019).
Rubbert-Roth, A. et al. Failure of anti-TNF treatment in patients with rheumatoid arthritis: the pros and cons of the early use of alternative biological agents. Autoimmun. Rev. 18, 102398 (2019).
Grivennikov, S. I. et al. Distinct and nonredundant in vivo functions of TNF produced by T cells and macrophages/neutrophils: protective and deleterious effects. Immunity 22, 93–104 (2005).
Bovensiepen, C. S. et al. TNF-producing Th1 cells are selectively expanded in liver infiltrates of patients with autoimmune hepatitis. J. Immunol. 203, 3148–3156 (2019).
Jain, A. et al. T cells instruct myeloid cells to produce inflammasome-independent IL-1β and cause autoimmunity. Nat. Immunol. 21, 65–74 (2020).
Schreurs, R. et al. Human fetal TNF-alpha-cytokine-producing CD4+ effector memory T cells promote intestinal development and mediate inflammation early in life. Immunity 50, 462–476.e468 (2019).
Weyand, C. M. & Goronzy, J. J. T-cell-targeted therapies in rheumatoid arthritis. Nat. Clin. Pr. Rheumatol. 2, 201–210 (2006).
Weyand, C. M. & Goronzy, J. J. Immunometabolism in early and late stages of rheumatoid arthritis. Nat. Rev. Rheumatol. 13, 291–301 (2017).
Slowikowski, K., Wei, K., Brenner, M. B. & Raychaudhuri, S. Functional genomics of stromal cells in chronic inflammatory diseases. Curr. Opin. Rheumatol. 30, 65–71 (2018).
Isaacs, J. D. Therapeutic T-cell manipulation in rheumatoid arthritis: past, present and future. Rheumatology 47, 1461–1468 (2008).
Probert, L. et al. Wasting, ischemia, and lymphoid abnormalities in mice expressing T cell-targeted human tumor necrosis factor transgenes. J. Immunol. 151, 1894–1906 (1993).
Weyand, C. M. & Goronzy, J. J. The immunology of rheumatoid arthritis. Nat. Immunol. 22, 10–18 (2021).
Yang, Z. et al. Restoring oxidant signaling suppresses proarthritogenic T cell effector functions in rheumatoid arthritis. Sci. Transl. Med. 8, 331ra338 (2016).
Yang, Z., Fujii, H., Mohan, S. V., Goronzy, J. J. & Weyand, C. M. Phosphofructokinase deficiency impairs ATP generation, autophagy, and redox balance in rheumatoid arthritis T cells. J. Exp. Med. 210, 2119–2134 (2013).
Wu, B. et al. Succinyl-CoA ligase deficiency in pro-inflammatory and tissue-invasive T cells. Cell Metab. 32, 967–980 e965 (2020).
Shen, Y. et al. Metabolic control of the scaffold protein TKS5 in tissue-invasive, proinflammatory T cells. Nat. Immunol. 18, 1025–1034 (2017).
Li, Y. et al. The DNA repair nuclease MRE11A functions as a aitochondrial protector and prevents T cell pyroptosis and tissue inflammation. Cell Metab. 30, 477–492.e476 (2019).
Wen, Z. et al. N-Myristoyltransferase deficiency impairs activation of kinase AMPK and promotes synovial tissue inflammation. Nat. Immunol. 20, 313–325 (2019).
Bommiasamy, H. et al. ATF6alpha induces XBP1-independent expansion of the endoplasmic reticulum. J. Cell Sci. 122, 1626–1636 (2009).
Sriburi, R., Jackowski, S., Mori, K. & Brewer, J. W. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J. Cell Biol. 167, 35–41 (2004).
Martinez-Reyes, I. & Chandel, N. S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 11, 102 (2020).
Zaslona, Z. & O’Neill, L. A. J. Cytokine-like roles for metabolites in immunity. Mol. Cell 78, 814–823 (2020).
Chinopoulos, C. Acute sources of mitochondrial NAD+ during respiratory chain dysfunction. Exp. Neurol. 327, 113218 (2020).
Korennykh, A. & Walter, P. Structural basis of the unfolded protein response. Annu. Rev. Cell Dev. Biol. 28, 251–277 (2012).
Jager, R., Bertrand, M. J., Gorman, A. M., Vandenabeele, P. & Samali, A. The unfolded protein response at the crossroads of cellular life and death during endoplasmic reticulum stress. Biol. Cell 104, 259–270 (2012).
Voorhees, R. M. & Hegde, R. S. Toward a structural understanding of co-translational protein translocation. Curr. Opin. Cell Biol. 41, 91–99 (2016).
Elvekrog, M. M. & Walter, P. Dynamics of co-translational protein targeting. Curr. Opin. Chem. Biol. 29, 79–86 (2015).
Schwarz, D. S. & Blower, M. D. The endoplasmic reticulum: structure, function and response to cellular signaling. Cell. Mol. Life Sci. 73, 79–94 (2016).
Sullivan, L. B. et al. Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell 162, 552–563 (2015).
Rubbert-Roth, A. et al. TNF inhibitors in rheumatoid arthritis and spondyloarthritis: are they the same? Autoimmun. Rev. 17, 24–28 (2018).
Li, Y. et al. Deficient activity of the nuclease MRE11A induces T cell aging and promotes arthritogenic effector functions in patients with rheumatoid arthritis. Immunity 45, 903–916 (2016).
Shaffer, A. L. et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21, 81–93 (2004).
Phillips, M. J. & Voeltz, G. K. Structure and function of ER membrane contact sites with other organelles. Nat. Rev. Mol. Cell Biol. 17, 69–82 (2016).
Hayashi, T. & Su, T. P. Sigma-1 receptor chaperones at the ER–mitochondrion interface regulate Ca2+ signaling and cell survival. Cell 131, 596–610 (2007).
Sebastian, D. et al. Mitofusin 2 (Mfn2) links mitochondrial and endoplasmic reticulum function with insulin signaling and is essential for normal glucose homeostasis. Proc. Natl Acad. Sci. USA 109, 5523–5528 (2012).
Simmen, T. et al. PACS-2 controls endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis. EMBO J. 24, 717–729 (2005).
Knupp, J., Arvan, P. & Chang, A. Increased mitochondrial respiration promotes survival from endoplasmic reticulum stress. Cell Death Differ. 26, 487–501 (2019).
Bravo, R. et al. Increased ER–mitochondrial coupling promotes mitochondrial respiration and bioenergetics during early phases of ER stress. J. Cell Sci. 124, 2143–2152 (2011).
Birsoy, K. et al. An essential role of the mitochondrial electron transport chain in cell proliferation Is to enable aspartate synthesis. Cell 162, 540–551 (2015).
Davila, A. et al. Nicotinamide adenine dinucleotide is transported into mammalian mitochondria. eLife https://doi.org/10.7554/eLife.33246 (2018).
Bettigole, S. E. & Glimcher, L. H. Endoplasmic reticulum stress in immunity. Annu. Rev. Immunol. 33, 107–138 (2015).
Gardner, B. M., Pincus, D., Gotthardt, K., Gallagher, C. M. & Walter, P. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb. Perspect. Biol. 5, a013169 (2013).
Chambers, J. E., Petrova, K., Tomba, G., Vendruscolo, M. & Ron, D. ADP ribosylation adapts an ER chaperone response to short-term fluctuations in unfolded protein load. J. Cell Biol. 198, 371–385 (2012).
Wu, B., Goronzy, J. J. & Weyand, C. M. Metabolic fitness of T cells in autoimmune disease. Immunometabolism https://doi.org/10.20900/immunometab20200017 (2020).
Kalliolias, G. D. & Ivashkiv, L. B. TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat. Rev. Rheumatol. 12, 49–62 (2016).
Yang, Z. et al. Restoring oxidant signaling suppresses proarthritogenic T cell effector functions in rheumatoid arthritis. Sci. Transl. Med. 8, 331ra338 (2016).
Li, Y. et al. Deficient activity of the nuclease MRE11A induces T cell aging and promotes arthritogenic effector functions in patients with rheumatoid arthritis. Immunity 45, 903–916 (2016).
Acknowledgements
This work was supported by the National Institutes of Health (grant nos. R01AR042527, R01AI108906, R01HL142068 and P01HL129941 to C.M.W. and nos. R01AI108891, R01AG045779, U19AI057266 and R01AI129191 to J.J.G.).
Author information
Authors and Affiliations
Contributions
C.M.W., J.J.G. and B.W. conceived the project. B.W., T.V.Z., Z.H. and K.J. formally analyzed and investigated the data. K.J.W. and M.P.A. recruited the patients. C.M.W., J.J.G. and B.W. wrote the original manuscript. C.M.W. and J.J.G. supervised the study. C.M.W. and J.J.G. acquired the funds.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Immunology thanks Navdeep Chandel and George Tsokos for their contribution to the peer review of this work. N. Bernard was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team. Peer review reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Mitochondrial mass in healthy and RA T cells.
CD4+CD45RA+ T cells were stimulated for 72 h. Flow cytometric quantification of mitochondria mass (MitoTrack Green MFI); n = 6 healthy and 6 RA. Data are mean ± SEM. Two-tailed unpaired Mann-Whitney-Wilcoxon rank test.
Extended Data Fig. 2 Gating strategy to analyze ER and mitochondrial function on the single cell level.
CD4+CD45RA+ T cells were stimulated for 72 h. ER biomass was determined with ER tracker and mitochondrial function was assessed with the mitochondrial membrane potential. Gate #1: small cellular size. Gate #2: medium cellular size. Gate #3: large cellular size.
Extended Data Fig. 3 XBP1S overexpression induces ER expansion.
Naive CD4+ T cells were stimulated and transfected with control or XBP1S overexpression plasmid before the ER size was determined. (a, b) Flow cytometry for ER Tracker staining; n = 4. (c) Confocal microscopy imaging of the ER protein calnexin. Scale bar, 10 μm, n = 3 independent experiments. (d, e) Flow cytometry for calnexin expression; n = 3. (f) XBP1S expression in T cells from patients treated with or w/o Methotrexate (MTX) (MTX: n = 13; w/o MTX: n = 12). All data are mean ± SEM. Two-tailed paired t test (b, e). Unpaired Mann-Whitney-Wilcoxon rank test (f). *P < 0.05, ***P < 0.001.
Extended Data Fig. 4 Inhibitors of mitochondrial respiration promote ER expansion.
Healthy naive CD4+ T cells were stimulated for 72 h in the presence of the mitochondrial respiration inhibitors Piericidin A (10 pM), Antimycin A (10 nM) or Oligomycin (1 nM). ER size was determined by flow cytometry measuring ER tracker (n = 4). Data are mean ± SEM. One-way ANOVA and post-ANOVA pair-wise two-group comparisons conducted with Tukey’s method. **P < 0.01.
Extended Data Fig. 5 Experimental scheme for mitochondria transfer.
(a) Experimental scheme for mitochondria transfer into RA T cells. (b) Experimental scheme for mitochondria transfer in Jurkat T cells. Mitochondria were labeled with MitoTrackerRed and isolated, then transferred into recipient cells. (c) Flow cytometric analysis of MitoTracker Red intensity after mitochondria transfer. Ratio indicates donor cell number/recipient cell number. (d) Representative confocal imaging of exogenous mitochondria transferred into Jurkat T cells, n = 3 independent experiments, scale bar, 20 μm.
Extended Data Fig. 6 Expression of GOT1 and GOT2 in healthy and RA T cells.
Peripheral blood CD4+CD45RA+ T cells from patients with RA and age-matched healthy individuals were isolated and stimulated for 72 h. mRNA levels of GOT1 and GOT2 were determined by qPCR. n = 4 in each group. All data are mean ± SEM. Two-tailed unpaired Mann-Whitney-Wilcoxon rank test.
Extended Data Fig. 7 Mitochondrial complex I inhibitor Piercidin A inhibits ADP-ribosylation of BiP.
ADP-ribosylation of BiP in healthy CD4+ T cells treated with or w/o Piercidin A (10 pM) for 24 h. n = 2.
Extended Data Fig. 8 Isolation of Rough ER.
(a) Naïve CD4+ T cells were purified from peripheral blood mononuclear cells and stimulated with anti-CD3/CD28 for 72 h. The rough ER was isolated by calcium precipitation and the isolate was immunoblotted for the ER protein calnexin, the ribosomal protein L17 and the cytoplasmic protein a-actin. (b) Healthy CD4+ T cells were activated with PMA/Ionomycin for 2 h before isolation of the rough ER and immunoblotting of the ER protein calnexin, the ribosomal protein S7 and the cytosolic protein β-actin.
Extended Data Fig. 9 Effects of Tunicamycin, aspartate, asparagine, pyruvate, and α-ketobutyrate on ER size and TNF production.
Naïve CD4+ T cells were purified from PBMCs and stimulated with anti-CD3/CD28 beads for 72 h in the presence of the indicated molecules. ER size quantified by MFI of ER tracker staining and TNF production measured by intracellular staining of TNF after PMA/ION stimulation for 2 h in the presence of the secretion inhibitor BFA. (a) Fold change of ER size after Tunicamycin treatment compared to the control group, n = 6. (b) Fold change of TNF production after Tunicamycin treatment compared to the control group, n = 4. (c) ER size change after Aspartate (1 mM) or Asparagine (1 mM) treatment, n = 6. (d) TNF production after aspartate and asparagine treatment, n = 3. (e) ER size change after pyruvate (1 mM) or α-KB (1 mM) treatment, n = 6. (f) TNF production after pyruvate or α-KB treatment, n = 4. All data are mean ± SEM, one-way ANOVA and post-ANOVA pair-wise two-group comparisons conducted with Tukey’s method. *P < 0.05, **P < 0.01, ***P < 0.001.
Extended Data Fig. 10 Mitochondria transfer into CD4+ T cells protects synovial tissue from inflammation.
Mitochondria were isolated from healthy T cells and transferred into RA CD4+ T cells prior to their adoptive transfer into synovium-NSG chimeras. Explanted synovial grafts were analyzed by immunohistochemical staining and tissue transcriptomics (RT-PCR). 8 tissues in each group. (a) H&E staining of synovial tissue sections. Scale bar; 50 μm. (b) Immunofluorescence staining for CD3+ T cells in synovial infiltrates. Representative images. Scale bar; 10 μm. (c) Gene expression profiling (RT-PCR) of TRB, TBET, RORG and other key inflammatory markers (n = 8). All data are mean ± SEM. Two-tailed unpaired Mann-Whitney-Wilcoxon rank test. *P < 0.05, **P < 0.01, ***P < 0.001.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2, and Supplementary Figs. 1–3.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data and unprocessed immunobots.
Source Data Fig. 6
Statistical source data and unprocessed immunobots.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data and unprocessed immunobots.
Source Data Extended Data Fig. 8
Unprocessed immunoblots.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
About this article
Cite this article
Wu, B., Zhao, T.V., Jin, K. et al. Mitochondrial aspartate regulates TNF biogenesis and autoimmune tissue inflammation. Nat Immunol 22, 1551–1562 (2021). https://doi.org/10.1038/s41590-021-01065-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-021-01065-2
This article is cited by
-
Lactobacillus paracasei L9 affects disease progression in experimental autoimmune neuritis by regulating intestinal flora structure and arginine metabolism
Journal of Neuroinflammation (2023)
-
Aging-associated HELIOS deficiency in naive CD4+ T cells alters chromatin remodeling and promotes effector cell responses
Nature Immunology (2023)
-
Amino acid metabolism in health and disease
Signal Transduction and Targeted Therapy (2023)
-
Efficient intervention for pulmonary fibrosis via mitochondrial transfer promoted by mitochondrial biogenesis
Nature Communications (2023)
-
Control of immune cell function by the unfolded protein response
Nature Reviews Immunology (2023)