Abstract
Blind mole rats (BMRs) are small rodents, characterized by an exceptionally long lifespan (>21 years) and resistance to both spontaneous and induced tumorigenesis. Here we report that cancer resistance in the BMR is mediated by retrotransposable elements (RTEs). Cells and tissues of BMRs express very low levels of DNA methyltransferase 1. Following cell hyperplasia, the BMR genome DNA loses methylation, resulting in the activation of RTEs. Upregulated RTEs form cytoplasmic RNA–DNA hybrids, which activate the cGAS–STING pathway to induce cell death. Although this mechanism is enhanced in the BMR, we show that it functions in mice and humans. We propose that RTEs were co-opted to serve as tumor suppressors that monitor cell proliferation and are activated in premalignant cells to trigger cell death via activation of the innate immune response. Activation of RTEs is a double-edged sword, serving as a tumor suppressor but contributing to aging in late life via the induction of sterile inflammation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Most data are included in the figures. The exact P values, if applicable, are included in the source data. RNA-seq and MeDIP data have been deposited in the Gene Expression Omnibus (GEO) under accession no. GSE181413. Normalized methylation values of the Illumina microarray (HorvathMammalMethylChip40) are available at the GEO under accession no. GSE181732. The RNA-seq data of 188 lung cancer cell lines were obtained from the Broad Institute (https://sites.broadinstitute.org/ccle/). The cell-line mutation information was obtained from http://amp.pharm.mssm.edu/Harmonizome/. Source data are provided with this paper.
Code availability
All codes used in the current study have been deposited at https://github.com/qwzhang0601/Transposon-triggered-innate-immune-response-confers-cancer-resistance-to-the-blind-mole-rat.
References
Seluanov, A., Gladyshev, V. N., Vijg, J. & Gorbunova, V. Mechanisms of cancer resistance in long-lived mammals. Nat. Rev. Cancer 18, 433–441 (2018).
Gorbunova, V. et al. Cancer resistance in the blind mole rat is mediated by concerted necrotic cell death mechanism. Proc. Natl Acad. Sci. USA 109, 19392–19396 (2012).
Manov, I. et al. Pronounced cancer resistance in a subterranean rodent, the blind mole-rat, Spalax: in vivo and in vitro evidence. BMC Biol. 11, 91 (2013).
Nevo, E. Mosaic Evolution of Subterranean Mammals: Regression, Progression, and Global Convergence (Oxford Univ. Press, 1999).
Bourque, G. et al. Ten things you should know about transposable elements. Genome Biol. 19, 199 (2018).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002).
Pace, J. K. 2nd & Feschotte, C. The evolutionary history of human DNA transposons: evidence for intense activity in the primate lineage. Genome Res. 17, 422–432 (2007).
Gasior, S. L., Wakeman, T. P., Xu, B. & Deininger, P. L. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 357, 1383–1393 (2006).
Iskow, R. C. et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 141, 1253–1261 (2010).
Gilbert, N., Lutz-Prigge, S. & Moran, J. V. Genomic deletions created upon LINE-1 retrotransposition. Cell 110, 315–325 (2002).
Slotkin, R. K. & Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272–285 (2007).
Leonova, K. I. et al. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc. Natl Acad. Sci. USA 110, E89-98 (2013).
Li, E., Bestor, T. H. & Jaenisch, R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926 (1992).
Robert, M. F. et al. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat. Genet. 33, 61–65 (2003).
De Cecco, M. et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566, 73–78 (2019).
Simon, M. et al. LINE1 derepression in aged wild-type and SIRT6-deficient mice drives inflammation. Cell Metab. 29, 871–885 (2019).
Fukuda, S. et al. Cytoplasmic synthesis of endogenous Alu complementary DNA via reverse transcription and implications in age-related macular degeneration. Proc. Natl Acad. Sci. USA 118, e2022751118 (2021).
Lee, E. et al. Landscape of somatic retrotransposition in human cancers. Science 337, 967–971 (2012).
Helman, E. et al. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res. 24, 1053–1063 (2014).
Tubio, J. M. C. et al. Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science 345, 1251343 (2014).
Miki, Y. et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 52, 643–645 (1992).
Burns, K. H. Our conflict with transposable elements and its implications for human disease. Annu Rev. Pathol. 15, 51–70 (2020).
Chiappinelli, K. B. et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162, 974–986 (2015).
Sheng, W. et al. LSD1 ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell 174, 549–563 (2018).
Gu, Z. et al. Silencing of LINE-1 retrotransposons is a selective dependency of myeloid leukemia. Nat. Genet. 53, 672–682 (2021).
Numajiri, A., Mibayashi, M. & Nagata, K. Stimulus-dependent and domain-dependent cell death acceleration by an IFN-inducible protein, human MxA. J. Interferon Cytokine Res. 26, 214–219 (2006).
Harada, H. et al. Absence of the type I IFN system in EC cells: transcriptional activator (IRF-1) and repressor (IRF-2) genes are developmentally regulated. Cell 63, 303–312 (1990).
Jin, J. et al. Noncanonical NF-κB pathway controls the production of type I interferons in antiviral innate immunity. Immunity 40, 342–354 (2014).
Hou, Y. et al. Non-canonical NF-κB antagonizes STING sensor-mediated DNA sensing in radiotherapy. Immunity 49, 490–503 (2018).
Dou, Z. et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550, 402–406 (2017).
Quentin, Y. A master sequence related to a free left Alu monomer (FLAM) at the origin of the B1 family in rodent genomes. Nucleic Acids Res. 22, 2222–2227 (1994).
Slack, A., Cervoni, N., Pinard, M. & Szyf, M. DNA methyltransferase is a downstream effector of cellular transformation triggered by simian virus 40 large T antigen. J. Biol. Chem. 274, 10105–10112 (1999).
Mankan, A. K. et al. Cytosolic RNA:DNA hybrids activate the cGAS–STING axis. EMBO J. 33, 2937–2946 (2014).
Dai, L., Huang, Q. & Boeke, J. D. Effect of reverse transcriptase inhibitors on LINE-1 and Ty1 reverse transcriptase activities and on LINE-1 retrotransposition. BMC Biochem. 12, 18 (2011).
Hahn, W. C. et al. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol. Cell. Biol. 22, 2111–2123 (2002).
Tian, X. et al. Evolution of telomere maintenance and tumour suppressor mechanisms across mammals. Philos. Trans. R Soc. B 373, 20160443 (2018).
Roulois, D. et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162, 961–973 (2015).
Zhang, W. & Xu, J. DNA methyltransferases and their roles in tumorigenesis. Biomark. Res. 5, 1 (2017).
Schaffer, B. S. et al. Immune competency of a hairless mouse strain for improved preclinical studies in genetically engineered mice. Mol. Cancer Ther. 9, 2354–2364 (2010).
Jin, Y., Tam, O. H., Paniagua, E. & Hammell, M. TEtranscripts: a package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics 31, 3593–3599 (2015).
Fang, X. et al. Genome-wide adaptive complexes to underground stresses in blind mole rats Spalax. Nat. Commun. 5, 3966 (2014).
Gibbs, R. A. et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428, 493–521 (2004).
Ashur-Fabian, O. et al. Evolution of p53 in hypoxia-stressed Spalax mimics human tumor mutation. Proc. Natl Acad. Sci. USA 101, 12236–12241 (2004).
Imbeault, M., Helleboid, P. Y. & Trono, D. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature 543, 550–554 (2017).
Schneider, K. et al. Dissection of cell cycle-dependent dynamics of Dnmt1 by FRAP and diffusion-coupled modeling. Nucleic Acids Res. 41, 4860–4876 (2013).
Mehdipour, P. et al. Epigenetic therapy induces transcription of inverted SINEs and ADAR1 dependency. Nature 588, 169–173 (2020).
Liu, H. et al. Tumor-derived IFN triggers chronic pathway agonism and sensitivity to ADAR loss. Nat. Med. 25, 95–102 (2019).
Leeansyah, E. et al. Inhibition of telomerase activity by human immunodeficiency virus (HIV) nucleos(t)ide reverse transcriptase inhibitors: a potential factor contributing to HIV-associated accelerated aging. J. Infect. Dis. 207, 1157–1165 (2013).
Song, L. et al. Nucleoside/nucleotide reverse transcriptase inhibitors attenuate angiogenesis and lymphangiogenesis by impairing receptor tyrosine kinases signalling in endothelial cells. Br. J. Pharm. 175, 1241–1259 (2018).
Barillari, G. The anti-angiogenic effects of anti-human immunodeficiency virus drugs. Front. Oncol. 10, 806 (2020).
Armando, R. G., Mengual Gomez, D. L. & Gomez, D. E. New drugs are not enough—drug repositioning in oncology: an update. Int. J. Oncol. 56, 651–684 (2020).
Ting, D. T. et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 331, 593–596 (2011).
Wylie, A. et al. p53 genes function to restrain mobile elements. Genes Dev. 30, 64–77 (2016).
Jangam, D., Feschotte, C. & Betran, E. Transposable element domestication as an adaptation to evolutionary conflicts. Trends Genet. 33, 817–831 (2017).
Matheu, A., Maraver, A. & Serrano, M. The Arf/p53 pathway in cancer and aging. Cancer Res. 68, 6031–6034 (2008).
Tyner, S. D. et al. p53 mutant mice that display early ageing-associated phenotypes. Nature 415, 45–53 (2002).
Coppe, J. P. et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 6, 2853–2868 (2008).
Campisi, J. & d’Adda di Fagagna, F. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8, 729–740 (2007).
Birney, E., Clamp, M. & Durbin, R. GeneWise and Genomewise. Genome Res. 14, 988–995 (2004).
Zhao, Y. et al. Naked mole rats can undergo developmental, oncogene-induced and DNA damage-induced cellular senescence. Proc. Natl Acad. Sci. USA 115, 1801–1806 (2018).
Franken, N. A., Rodermond, H. M., Stap, J., Haveman, J. & van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 1, 2315–2319 (2006).
Debacq-Chainiaux, F., Erusalimsky, J. D., Campisi, J. & Toussaint, O. Protocols to detect senescence-associated beta-galactosidase (SA-βgal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc. 4, 1798–1806 (2009).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Tarailo-Graovac, M. & Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. Chapter 4, Unit 4.10 (2009).
Bao, W., Kojima, K. K. & Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Yates, A. D. et al. Ensembl 2020. Nucleic Acids Res. 48, D682–D688 (2020).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Tian, X. et al. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature 499, 346–349 (2013).
Klijn, C. et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 33, 306–312 (2015).
Anandakrishnan, R., Varghese, R. T., Kinney, N. A. & Garner, H. R. Estimating the number of genetic mutations (hits) required for carcinogenesis based on the distribution of somatic mutations. PLoS Comput. Biol. 15, e1006881 (2019).
Acknowledgements
This research was supported by grants from the US National Institutes of Health to Z. Zhang (grant no. AG047200), A.S. (grant no. AG047200) and V.G. (grant nos AG047200 and AG051449).
Author information
Authors and Affiliations
Contributions
Y.Z., M.S., S.H., E.N., A.S. and V.G. designed the research. Y.Z. performed most of the experiments. E.O. performed the experiments with DNMT1 expression. J.L. performed the DNA-damage experiments. Q.Z., J.Y.L., Z. Zheng and Z. Zhang analyzed data. Z.K. and J.A. performed the DMBA–TPA experiments. Q.L., A.G., Y.S.L., J.H., J.L. and Z. Zheng provided help with experiments. A.S. and V.G. supervised the research. Y.Z., A.S. and V.G. wrote the paper with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Immunology thanks the anonymous reviewers for their contribution to the peer review of this work. Jamie D. K. Wilson was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Activation of SASP in overproliferating BMR cells and differential gene expression in BMR and mice tissues.
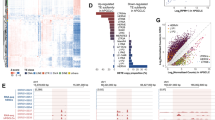
(a) qRT-PCR showing senescence markers P21 and P16 were increased with proliferation of BMR cells. (b) SASP factors IL1b, IL6, IL8, and MMP12 were increased with proliferation of BMR cells. (c) KEGG pathway enrichment analysis of consistently upregulated and downregulated genes in the BMR tissues. RNA samples from 8 different tissues, including intestine, kidney, liver, lung, muscle, skin, spleen, and white adipose tissues (WAT) of BMR and mice were sequenced. A gene is defined as consistently upregulated gene if it expresses higher in BMR than in mice in at least six tissues. P values were adjusted by FDR. (d) A boxplot showing different fold changes of genes from IFN and NF-κB pathways comparing BMR and mice tissues. Center lines represent median, bounds of box are the 1st quartile and 3rd quartile, and whiskers are drawn down to the 10th percentile and up to the 90th percentile. (e) A heatmap showing differential gene expression in BMR CCD cells.
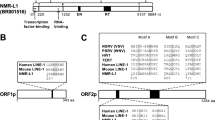
Extended Data Fig. 2 Transcriptional start sites of B1 and L1 in BMR CCD cells.
(a) Transcriptional start sites of B1 and L1 in CCD cells of BMR. 5′ RACE was performed using BMR fibroblasts undergoing CCD. The RACE product was ligated into T vector and colonies were sequenced. A multiple alignment of the products was generated against the consensus SINE B1 sequence or a predicted active L1 family homologous to mouse L1MdaI, respectively. The alignment was generated with Clustal Omega and displayed by Jalview. The location of gene-specific primer 2 (GSP2) for both B1 and L1 were shown under the consensus sequence. (b) Summary of the mapping data of L1 5′ RACE. The location of transcription start sites were shown relative to the consensus start site.
Extended Data Fig. 3 RNA expression of SINE B1, B2 and IFNB1 in carcinogen DMBA/TPA treated BMR and mouse skin.
BMR and mice were topically treated with DMBA followed by treatment of TPA 3 days after DMBA. Two weeks after treatment, biopsy was taken from treated area, and RNA was extracted. Expression of SINE B1, B2 and IFNB1 was determined by qRT-PCR. Data are mean ± SEM of 3 biological replicates of 2 technical repeats.
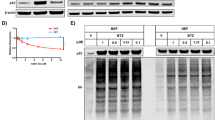
Extended Data Fig. 4 Naturally low levels of DNMT1 result in loss of DNA methylation and SASP in overproliferating BMR cells.
(a) Unsupervised hierarchical clustering of DNA samples from BMR and mouse fibroblasts with low and high PDs. The Average linkage hierarchical clustering is based on the inter-array correlation coefficient (Pearson correlation). The cluster branches (first color band) correspond to species (second color band, blue = BMR, cyan = mouse). Third color band: high PD (O; black) versus low PD (Y; white). (b) A heatmap showing differentially methylated elements determined using Methylated DNA immunoprecipitation (MeDIP). Differentially methylated regions (DMRs) were annotated against BMR genome. Totally 39 TE families were hyper-methylated and 60 TE families were hypo-methylated in dying BMR cells. Hypo-methylated TEs are enriched with B1 (especially PB1D9 family) and L1. (c) A heatmap showing generally lower expression levels of DNMT1 in 8 different tissues of BMR than mice. (d) alignment of partial sequence of human, mouse and BMR DNMT1 protein including immunogen recognized by the antibody used (Abcam ab13537). The immunogen is highly conserved across species, which is identical between human and BMR, and only 3 different amino acids different in mouse DNMT1. (e) Western blot detecting endogenous DNMT1 of primary and Large T transformed BMR cells. Large T is known to elevate DNMT1 expression. The significant band shows that the antibody is capable to detect BMR DNMT1. The experiment was repeated three times independently with similar results. (f) qRT-PCR showing overexpression of human DNMT1 in BMR cells. (g) Overexpression of DNMT1 restored global DNA methylation in BMR cells. (h) qRT-PCR showing repressed senescence factors by overexpression of DNMT1. Data are mean ± SD of 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 by unpaired two-sided Student’s t-test.
Extended Data Fig. 5 BMR cells undergo DNA damage induced by IFN.
(a) Images and quantification of comet assay detecting DNA damage of BMR cells with proliferation. Scale bar, 10 μM. (b) Images and quantification of immunofluorescence detecting γH2AX foci of BMR cells with proliferation. Scale bar, 5 μM. (c) DNA damage response of BMR cells with proliferation. Western blot was performed to show the phosphorylation of Chk2 (Thr68) and p53 (Ser15). The experiment was repeated three times independently with similar results. (d) Comet assay showing reduced DNA damage by overexpression of DNMT1. Scale bar, 10 μM. (e) Comet assay showing DNA damage induced by IFN. Young, growing BMR cells were treated with either medium containing BMR IFN or conditioned medium from old, dying BMR cells for 12 days before harvesting for comet assay. Scale bar, 10 μM. Tail DNA percentage (a, d, and e) or γH2AX (b) foci were quantified from 100 cells per group. Data are mean ± . **P < 0.01, ***P < 0.001 by unpaired two-sided Student’s t-test.
Extended Data Fig. 6 Nucleoside reverse-transcriptase inhibitors (NRTIs) treatment reduces RNA/DNA hybrids and represses cGAS–STING pathway.
(a) Images of cytoplasmic double-stranded (ds) RNA in young and dying BMR cells stained with J2 antibody. Cells treated with RNase III and with secondary antibody only lost the signals, confirming the specificity of J2 antibody recognizing dsRNA. Scale bar, 5 μM. The experiment was repeated three times independently with similar results. (b) Images of cytoplasmic RNA/DNA hybrids in BMR cells stained with S9.6 antibody. Treatment of RNase H, which specifically degrades RNA/DNA hybrids, and with secondary antibody only didn’t yield signals, confirming that S9.6 antibody specifically recognizes RNA/DNA hybrids. Scale bar, 5 μM. The experiment was repeated three times independently with similar results. (c) Reduced cytoplasmic RNA/DNA hybrids by NRTIs treatment stained by S9.6 antibody in BMR cells. Scale bar, 5 μM. Immunofluorescence intensities were quantified from 30 cells per group. (d) Comet assay showing reduced DNA damage by NRTIs treatment in BMR cells. Scale bar, 10 μM. Tail DNA percentages were quantified from 100 cells per group. (e-f) knockdown of cGAS (e) and STING (f) were determined by qRT-PCR and western blot. (g) growth curve showing that knockdown of dsRNA-sensing genes TLR3, MAVS, MDA-5, and IRG-1 didn’t rescue CCD. Data are mean ± SD of 3 independent experiments. **P < 0.01, ***P < 0.001 by unpaired two-sided Student’s t-test.
Extended Data Fig. 7 Roles of DNMT1 and RTEs in tumorigenesis in mice and human cells.
(a) Knockdown of DNMT1 in HeLa and HT1080 cancer cells as determined by Western blot. The experiment was repeated three times independently with similar results. (b-c) Knockdown of DNMT1 repressed proliferation of HT1080 cells in vitro. Clonogenic assay was performed with crystal violet staining and colonies areas were quantified (c). N = 3 technical replicates. Experiment was repeated three times independently with similar results. (d) Growth curve of DMBA/TPA-induced papilloma in mice treated with or without abacavir. Abacavir treated alone repressed DMBA/TPA induced papilloma. Experiment was performed using eight animals for each group. (e) Images of SKH1 hairless mice with papilloma induced by DMBA/TPA. 5-Aza repressed formation of papilloma, while abacavir restored this repression. (f) qRT-PCR showing overexpression of SINE B1 in HeLa cells. N = 3 technical replicates. (g) Western blot showing overexpression of L1 (ORFeus) in HeLa cells by detecting ORF1p. The experiment was repeated three times independently with similar results. (h) qRT-PCR showing activation of IFNB1 in HeLa cells by overexpression of SINE B1 and L1 (ORFeus). N = 3 technical replicates. Data are mean ± SD (c, f, and h) or mean ± SEM (d). * P < 0.05, **P < 0.01, ***P < 0.001 by unpaired two-sided Student’s t-test.
Supplementary information
Supplementary Data 1
IFN-pathway gene list.
Supplementary Data 2
CCLE accession number and web links.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 6
Unprocessed western blots.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 7
Unprocessed western blots.
Rights and permissions
About this article
Cite this article
Zhao, Y., Oreskovic, E., Zhang, Q. et al. Transposon-triggered innate immune response confers cancer resistance to the blind mole rat. Nat Immunol 22, 1219–1230 (2021). https://doi.org/10.1038/s41590-021-01027-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-021-01027-8
This article is cited by
-
A zinc finger transcription factor enables social behaviors while controlling transposable elements and immune response in prefrontal cortex
Translational Psychiatry (2024)
-
Towards targeting transposable elements for cancer therapy
Nature Reviews Cancer (2024)
-
Cancer-derived exosomal Alu RNA promotes colorectal cancer progression
Experimental & Molecular Medicine (2024)
-
Regulation and function of transposable elements in cancer genomes
Cellular and Molecular Life Sciences (2024)
-
DNA damage and repair in age-related inflammation
Nature Reviews Immunology (2023)