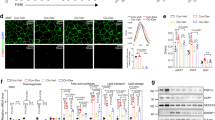
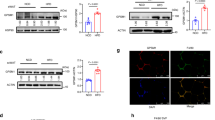
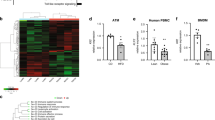
Abstract
Metabolic inflammation is closely linked to obesity, and is implicated in the pathogenesis of metabolic diseases. FTO harbors the strongest genetic association with polygenic obesity, and IRX3 mediates the effects of FTO on body weight. However, in what cells and how IRX3 carries out this control are poorly understood. Here we report that macrophage IRX3 promotes metabolic inflammation to accelerate the development of obesity and type 2 diabetes. Mice with myeloid-specific deletion of Irx3 were protected against diet-induced obesity and metabolic diseases via increasing adaptive thermogenesis. Mechanistically, macrophage IRX3 promoted proinflammatory cytokine transcription and thus repressed adipocyte adrenergic signaling, thereby inhibiting lipolysis and thermogenesis. JNK1/2 phosphorylated IRX3, leading to its dimerization and nuclear translocation for transcription. Further, lipopolysaccharide stimulation stabilized IRX3 by inhibiting its ubiquitination, which amplified the transcriptional capacity of IRX3. Together, our findings identify a new player, macrophage IRX3, in the control of body weight and metabolic inflammation, implicating IRX3 as a therapeutic target.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Sequencing data generated for this study have been deposited in the Gene Expression Omnibus database under accession number GSE178574. Source data are provided with this paper.
References
Lowell, B. B. & Spiegelman, B. M. Towards a molecular understanding of adaptive thermogenesis. Nature 404, 652–660 (2000).
Wu, J. et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 150, 366–376 (2012).
Wang, W. & Seale, P. Control of brown and beige fat development. Nat. Rev. Mol. Cell Biol. 17, 691–702 (2016).
Qiu, Y., Shan, B., Yang, L. & Liu, Y. Adipose tissue macrophage in immune regulation of metabolism. Sci. China Life Sci. 59, 1232–1240 (2016).
Harms, M. & Seale, P. Brown and beige fat: development, function and therapeutic potential. Nat. Med. 19, 1252–1263 (2013).
Rosen, E. D. & Spiegelman, B. M. What we talk about when we talk about fat. Cell 156, 20–44 (2014).
Mathis, D. Immunological goings-on in visceral adipose tissue. Cell Metab. 17, 851–859 (2013).
Mowers, J. et al. Inflammation produces catecholamine resistance in obesity via activation of PDE3B by the protein kinases IKKε and TBK1. Elife 2, e01119 (2013).
Sakamoto, T. et al. Macrophage infiltration into obese adipose tissues suppresses the induction of UCP1 level in mice. Am. J. Physiol. Endocrinol. Metab. 310, E676–E687 (2016).
Chawla, A., Nguyen, K. D. & Goh, Y. P. Macrophage-mediated inflammation in metabolic disease. Nat. Rev. Immunol. 11, 738–749 (2011).
Dina, C. et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 39, 724–726 (2007).
Frayling, T. M. et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316, 889–894 (2007).
Scuteri, A. et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 3, e115 (2007).
Fischer, J. et al. Inactivation of the Fto gene protects from obesity. Nature 458, 894–898 (2009).
Church, C. et al. Overexpression of Fto leads to increased food intake and results in obesity. Nat. Genet. 42, 1086–1092 (2010).
Grunnet, L. G. et al. Regulation and function of FTO mRNA expression in human skeletal muscle and subcutaneous adipose tissue. Diabetes 58, 2402–2408 (2009).
Kloting, N. et al. Inverse relationship between obesity and FTO gene expression in visceral adipose tissue in humans. Diabetologia 51, 641–647 (2008).
Wahlen, K., Sjolin, E. & Hoffstedt, J. The common rs9939609 gene variant of the fat mass- and obesity-associated gene FTO is related to fat cell lipolysis. J. Lipid Res. 49, 607–611 (2008).
Smemo, S. et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507, 371–375 (2014).
Kim, K. H., Rosen, A., Bruneau, B. G., Hui, C. C. & Backx, P. H. Iroquois homeodomain transcription factors in heart development and function. Circ. Res. 110, 1513–1524 (2012).
de Araujo, T. M. & Velloso, L. A. Hypothalamic IRX3: a new player in the development of obesity. Trends Endocrinol. Metab. 31, 368–377 (2020).
Claussnitzer, M. et al. FTO obesity variant circuitry and adipocyte browning in humans. N. Engl. J. Med. 373, 895–907 (2015).
Zou, Y. et al. IRX3 promotes the browning of white adipocytes and its rare variants are associated with human obesity risk. EBioMedicine 24, 64–75 (2017).
de Araujo, T. M. et al. The partial inhibition of hypothalamic IRX3 exacerbates obesity. EBioMedicine 39, 448–460 (2019).
Cannon, B. & Nedergaard, J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 84, 277–359 (2004).
Morrison, S. F., Madden, C. J. & Tupone, D. Central neural regulation of brown adipose tissue thermogenesis and energy expenditure. Cell Metab. 19, 741–756 (2014).
Hoffmann, C., Leitz, M. R., Oberdorf-Maass, S., Lohse, M. J. & Klotz, K. N. Comparative pharmacology of human beta-adrenergic receptor subtypes–characterization of stably transfected receptors in CHO cells. Naunyn Schmiedebergs Arch. Pharmacol. 369, 151–159 (2004).
Duncan, R. E., Ahmadian, M., Jaworski, K., Sarkadi-Nagy, E. & Sul, H. S. Regulation of lipolysis in adipocytes. Annu. Rev. Nutr. 27, 79–101 (2007).
Fredriksson, J. M. et al. Analysis of inhibition by H89 of UCP1 gene expression and thermogenesis indicates protein kinase A mediation of β3-adrenergic signalling rather than β3-adrenoceptor antagonism by H89. Biochim. Biophys. Acta 1538, 206–217 (2001).
Brito, N. A., Brito, M. N. & Bartness, T. J. Differential sympathetic drive to adipose tissues after food deprivation, cold exposure or glucoprivation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 294, R1445–1452 (2008).
Qiu, Y. et al. Eosinophils and type 2 cytokine signaling in macrophages orchestrate development of functional beige fat. Cell 157, 1292–1308 (2014).
Goto, T. et al. Proinflammatory cytokine interleukin-1β suppresses cold-induced thermogenesis in adipocytes. Cytokine 77, 107–114 (2016).
Chung, K. J. et al. A self-sustained loop of inflammation-driven inhibition of beige adipogenesis in obesity. Nat. Immunol. 18, 654–664 (2017).
Huang, C. H. et al. UbiSite: incorporating two-layered machine learning method with substrate motifs to predict ubiquitin-conjugation site on lysines. BMC Syst. Biol. 10, 6 (2016).
Lu, Y. C., Yeh, W. C. & Ohashi, P. S. LPS/TLR4 signal transduction pathway. Cytokine 42, 145–151 (2008).
Lei, K. et al. The Bax subfamily of Bcl2-related proteins is essential for apoptotic signal transduction by c-Jun NH2-terminal kinase. Mol. Cell. Biol. 22, 4929–4942 (2002).
Han, M. S. et al. JNK expression by macrophages promotes obesity-induced insulin resistance and inflammation. Science 339, 218–222 (2013).
Wang, C. et al. GPS 5.0: an update on the prediction of kinase-specific phosphorylation sites in proteins. Genomics Proteomics Bioinformatics 18, 72–80 (2020).
Bilioni, A., Craig, G., Hill, C. & McNeill, H. Iroquois transcription factors recognize a unique motif to mediate transcriptional repression in vivo. Proc. Natl Acad. Sci. USA 102, 14671–14676 (2005).
Berger, M. F. et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell 133, 1266–1276 (2008).
Honda, K. & Taniguchi, T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 6, 644–658 (2006).
Villarino, A. V., Kanno, Y. & O’Shea, J. J. Mechanisms and consequences of Jak–STAT signaling in the immune system. Nat. Immunol. 18, 374–384 (2017).
Hill, D. A. et al. Distinct macrophage populations direct inflammatory versus physiological changes in adipose tissue. Proc. Natl Acad. Sci. USA 115, E5096–E5105 (2018).
Abram, C. L., Roberge, G. L., Hu, Y. & Lowell, C. A. Comparative analysis of the efficiency and specificity of myeloid-Cre deleting strains using ROSA-EYFP reporter mice. J. Immunol. Methods 408, 89–100 (2014).
Passegue, E., Wagner, E. F. & Weissman, I. L. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell 119, 431–443 (2004).
Orthgiess, J. et al. Neurons exhibit Lyz2 promoter activity in vivo: implications for using LysM-Cre mice in myeloid cell research. Eur. J. Immunol. 46, 1529–1532 (2016).
Gomez-Skarmeta, J. L., Diez del Corral, R., de la Calle-Mustienes, E., Ferre-Marco, D. & Modolell, J. Araucan and caupolican, two members of the novel iroquois complex, encode homeoproteins that control proneural and vein-forming genes. Cell 85, 95–105 (1996).
Creely, S. J. et al. Lipopolysaccharide activates an innate immune system response in human adipose tissue in obesity and type 2 diabetes. Am. J. Physiol. Endocrinol. Metab. 292, E740–747 (2007).
Lancaster, G. I. et al. Evidence that TLR4 is not a receptor for saturated fatty acids but mediates lipid-induced inflammation by reprogramming macrophage metabolism. Cell Metab. 27, 1096–1110 e1095 (2018).
Zhi, X. et al. AdipoCount: a new software for automatic adipocyte counting. Front Physiol. 9, 85 (2018).
Qiu, Y. et al. Eosinophils and type 2 cytokine signaling in macrophages orchestrate development of functional beige fat. Cell 157, 1292–1308 (2014).
Shan, B. et al. The metabolic ER stress sensor IRE1α suppresses alternative activation of macrophages and impairs energy expenditure in obesity. Nat. Immunol. 18, 519–529 (2017).
Acknowledgements
We thank members of the laboratory of Y.Q. and Y. Liu (Wuhan University) for helpful comments on the manuscript. We thank C.-C. Hui (University of Toronto) for generous sharing of Irx3fl/fl, Irx3−/− and Irx5−/− mice and mouse Irx3 cDNA. We thank Z. Dong (Tsinghua University) for generous sharing of CD1d tetramer (loaded with PBS-57) from the National Institutes of Health Tetramer Core Facility. We also thank H. Deng and X. Meng in Proteinomics Facility at Technology Center for Protein Sciences, Tsinghua University, for protein MS analysis. This work was supported by grants from National Key R&D Program of China (2018YFA0800702), National Natural Science Foundation of China (31671227 and 91642113) and the Thousand Young Talents Program of the Chinese government (to Y.Q.).
Author information
Authors and Affiliations
Contributions
J.Y., D.W. and C.Z. designed and performed the main experiments with assistance from T.Y., Y.Z., H.S., K.X., X.H. and Z.W.; J.Y., D.W., C.Z. and Y.Q. discussed and interpreted the results from the study; and J.Y., D.W. and Y.Q. conceived of the study, supervised the work and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Immunology thanks Andrew Hogan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. L. A. Dempsey was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 IRX3 deficiency in macrophages decreases fat accumulation, while does not affect food intake and activity in HFD-fed and ND-fed mice.
a-d, Representative images of H&E-stained sections of different tissues (a), and distribution of adipocyte size of BAT (b), scWAT (c) and eWAT (d) from Irx3f/f and Irx3f/fLyz2Cre mice fed HFD (n = 4). e-h, Representative images of H&E-stained sections of different tissues (e), and distribution of adipocyte size of BAT (f), scWAT (g) and eWAT (h) from Irx3f/f and Irx3f/fLyz2Cre mice fed ND (n = 3). i, j, Locomotor activity and food intake of Irx3f/f and Irx3f/fLyz2Cre mice fed HFD (i) (n = 10 for Irx3f/f, n = 9 for Irx3f/fLyz2Cre mice in locomotor activity and n = 4 in food intake) and ND (j) (n = 8 for Irx3f/f, n = 9 for Irx3f/fLyz2Cre mice in locomotor activity and n = 9 in food intake). All data represent means ± s.e.m. Statistical significance was determined by unpaired two-tailed Student’s t-test (b-d, f-h, i, j).
Extended Data Fig. 2 IRX3 deficiency in macrophages increases adipose tissues lipolysis.
a, b, Locomotor activity (a) and food intake (b) of Irx3f/f and Irx3f/fLyz2Cre mice housed at room temperature (22 °C) and cold environment (4 °C) (n = 6). c-e,Quantitative RT-PCR analysis of lipogenic gene expression in BAT (c), scWAT (d) and eWAT (e) of Irx3f/f and Irx3f/fLyz2Cre mice housed at 4 °C for 48 h (n = 6 for Irx3f/f and n = 5 for Irx3f/fLyz2Cre mice). f, RER from mice in Fig. 2a (n = 6). g, Quantitative RT-PCR analysis of thermogenic gene expression in eWAT of Irx3f/f and Irx3f/fLyz2Cre mice housed at 4 °C for 48 h (n = 5 or 6 for Irx3f/f and n = 5 or 6 for Irx3f/fLyz2Cre mice). h, i, NE-induced RER from mice in Fig. 3a (n = 5). All data represent means ± s.e.m. Statistical significance was determined by two-way ANOVA with Bonferroni’s multiple-comparisons test (f) or unpaired two-tailed Student’s t-test (a-e and g-i).
Extended Data Fig. 3 IRX3 promotes proinflammatory cytokine expression in macrophages to inhibit adipocyte lipolysis and thermogenesis.
a, Serum concentrations of adrenaline, noradrenaline and dopamine of Irx3f/f and Irx3f/fLyz2Cre mice housed at room temperature (22 °C) or 4 °C for 48 h (n = 5). b, c, Quantitative RT-PCR analysis of macrophage marker expression in BAT (b) and scWAT (c) of Irx3f/f and Irx3f/fLyz2Cre mice housed at 4 °C for 48 h (n = 6 for Irx3f/f and n = 4 or 5 for Irx3f/fLyz2Cre mice). d, Flow cytometry analysis of macrophage numbers in scWAT of Irx3f/f and Irx3f/fLyz2Cre mice housed at 4 °C for 48 h (n = 5). e, Quantitative RT-PCR analysis of M2 marker expression in scWAT of Irx3f/f and Irx3f/fLyz2Cre mice housed at 4 °C for 48 h (n = 6 for Irx3f/f and n = 5 or 6 for Irx3f/fLyz2Cre mice). f, g, Quantitative RT-PCR analysis of proinflammatory gene expression in BAT (f) and eWAT (g) of Irx3f/f and Irx3f/fLyz2Cre mice housed at 4 °C for 48 h (n = 6 for Irx3f/f and n = 5 for Irx3f/fLyz2Cre mice). h, Quantitative RT-PCR analysis of proinflammatory gene expression in peritoneal macrophage of Irx3f/f and Irx3f/fLyz2Cre mice (n = 3). i, Immunoblot analysis of beige adipocytes pretreated with 5 ng/ml cytokines and followed by a co-treatment of 50 nM NE for 8 hrs. Independent experiments were performed three times with similar results. j, k, Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of genes significantly upregulated (log2FC > 0.5, Padj<0.05). All data represent means ± s.e.m. Statistical significance was determined by unpaired two-tailed Student’s t-test (a-h).
Extended Data Fig. 4 LPS stimulation inhibits IRX3’s degradation, while increases its phosphorylation.
a, Immunoblot analysis of RAW 264.7 cells overexpressing IRX3-Flag and then treated with LPS for 12 h followed by CHX treatment for indicated time, and IRX3 protein level was quantified by ImageJ. b, Ubiquitination site prediction by ‘Ubisite’. c, Immunoblot analysis of RAW 264.7 cells overexpressing IRX3 (WT) and its ubiquitination site mutants and then treated with CHX for indicated time, and quantification of IRX3 protein level by ImageJ. d, Immunoblot analysis and phos-tag SDS-PAGE analysis of RAW 264.7 cells overexpressing IRX3-HA plus indicated kinases in RAW 264.7 cells and then treated with or without LPS for 4 h. Independent experiments were performed three times with similar results. e, JNK phosphorylation site prediction by ‘GPS 5.0’. f, Alignment of IRX3 sequences in different species. IP-MS-identified Ser phosphorylation sites (S361 and S389) were highlighted in red. g, h, Quantitative RT-PCR analysis of proinflammatory gene expression in human THP-1 monocytes overexpressing GFP or hIRX3 (g) or in Cas9 stable THP-1 monocytes infected with control or hIRX3-gRNA AAVs (h) (n = 3). (i-l) Quantitative RT-PCR analysis of proinflammatory gene expression in differentiated human THP-1 macrophages overexpressing hIRX3 (i) or hIRX3-shRNA (j), or in hMDMs overexpressing hIRX3 (k) or hIRX3-shRNA (l) (n = 3). Statistical significance was determined by unpaired two-tailed Student’s t-test (g-l).
Extended Data Fig. 5 Profiling of crown-like structure and different immune cell populations in Irx3f/f and Irx3f/fLyz2Cre mice scWAT.
a, b, Representative images of F4/80 IHC staining (a) and quantification of CLS (b) in scWAT from Irx3f/f and Irx3f/fLyz2Cre mice fed HFD (n = 6). c, d, Gating strategy for analysis of different subpopulations of macrophages (c) and other immune cells (d) in scWAT of Irx3f/f and Irx3f/fLyz2Cre mice fed HFD. e-i, Flow cytometry analysis of different subpopulations of macrophages (e), T cells, B cells (f), NK cells (g), CD4+ T cells, CD8+ T cells, Treg cells (h) and NKT cells (i) (n = 4 in e, n = 5 for Irx3f/f and n = 6 for Irx3f/fLyz2Cre mice in f, h and n = 6 in g, i). All data represent means ± s.e.m. Statistical significance was determined by unpaired two-tailed Student’s t-test (b, e-i).
Extended Data Fig. 6 IRX3 depletion in neutrophils does not affect the HFD-induced metabolic dysfunctions.
a-d, Body weight (a), tissues weight (b), GTT (c) and ITT (d) of Irx3f/f and Irx3f/fS100a8Cre mice fed HFD (n = 6). e, In situ hybridization of Irx3 RNA (red) in conjunction with staining for the neuronal marker NeuN (green) and DAPI (Blue) in the sagittal brain sections of Irx3f/f and Irx3f/fLyz2Cre mice. f, Quantitative RT-PCR analysis of Irx3 expression in the whole brains and hypothalami of Irx3f/f and Irx3f/fLyz2Cre mice (n = 4). g, Quantitative RT-PCR analysis of proinflammatory gene expression in WT and Irx5−/− BMDM (n = 3). h, Luciferase assay of Il1a, Il1b and Il6 promoter-driven luciferase reporter in HEK 293 T cells co-transfected with GFP or IRX5 plasmid for 24 h (n = 3). i, Model of how macrophage IRX3 controls proinflammatory cytokine expression and thus inhibits adipocyte thermogenesis and lipolysis. All data represent means ± s.e.m. Statistical significance was determined by two-way ANOVA with Bonferroni’s multiple-comparisons test (a, c and d) or unpaired two-tailed Student’s t-test (b, f-h).
Supplementary information
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 6
Unprocessed western blots.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 7
Unprocessed western blots.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
About this article
Cite this article
Yao, J., Wu, D., Zhang, C. et al. Macrophage IRX3 promotes diet-induced obesity and metabolic inflammation. Nat Immunol 22, 1268–1279 (2021). https://doi.org/10.1038/s41590-021-01023-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-021-01023-y
This article is cited by
-
METTL3 and METTL14-mediated N6-methyladenosine modification of SREBF2-AS1 facilitates hepatocellular carcinoma progression and sorafenib resistance through DNA demethylation of SREBF2
Scientific Reports (2024)
-
Mapping novel QTL and fine mapping of previously identified QTL associated with glucose tolerance using the collaborative cross mice
Mammalian Genome (2024)
-
Apolipoprotein E (ApoE) orchestrates adipose tissue inflammation and metabolic disorders through NLRP3 inflammasome
Molecular Biomedicine (2023)
-
Transcriptional repression of beige fat innervation via a YAP/TAZ-S100B axis
Nature Communications (2023)
-
N6-methyladenosine of Spi2a attenuates inflammation and sepsis-associated myocardial dysfunction in mice
Nature Communications (2023)