Abstract
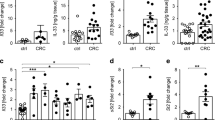
The transcription factor TCF-1 is essential for the development and function of regulatory T (Treg) cells; however, its function is poorly understood. Here, we show that TCF-1 primarily suppresses transcription of genes that are co-bound by Foxp3. Single-cell RNA-sequencing analysis identified effector memory T cells and central memory Treg cells with differential expression of Klf2 and memory and activation markers. TCF-1 deficiency did not change the core Treg cell transcriptional signature, but promoted alternative signaling pathways whereby Treg cells became activated and gained gut-homing properties and characteristics of the TH17 subset of helper T cells. TCF-1-deficient Treg cells strongly suppressed T cell proliferation and cytotoxicity, but were compromised in controlling CD4+ T cell polarization and inflammation. In mice with polyposis, Treg cell–specific TCF-1 deficiency promoted tumor growth. Consistently, tumor-infiltrating Treg cells of patients with colorectal cancer showed lower TCF-1 expression and increased TH17 expression signatures compared to adjacent normal tissue and circulating T cells. Thus, Treg cell–specific TCF-1 expression differentially regulates TH17-mediated inflammation and T cell cytotoxicity, and can determine colorectal cancer outcome.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The bulk RNA-seq and scRNA-seq datasets are deposited in the Gene Expression Omnibus under accession GSE163084. Source data are provided with this paper.
Code availability
The codes used for bulk RNA-seq and scRNA-seq analysis followed typical pipelines from public R packages (DESeq2 and Seurat). All codes are available upon request.
References
Fontenot, J. D., Gavin, M. A. & Rudensky, A. Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4, 330–336 (2003).
Hori, S., Nomura, T. & Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003).
Khattri, R., Cox, T., Yasayko, S. A. & Ramsdell, F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat. Immunol. 4, 337–342 (2003).
Benoist, C. & Mathis, D. Treg cells, life history and diversity. Cold Spring Harb. Perspect. Biol. 4, a007021 (2012).
Ohnmacht, C. et al. The microbiota regulates type 2 immunity through RORγt+ T cells. Science 349, 989–993 (2015).
Schiering, C. et al. The alarmin IL-33 promotes regulatory T cell function in the intestine. Nature 513, 564–568 (2014).
Pratama, A., Schnell, A., Mathis, D. & Benoist, C. Developmental and cellular age direct conversion of CD4+T cells into RORγ+ or Helios+ colon Treg cells.J. Exp. Med. 217, e20190428 (2020).
Zhou, J., Nefedova, Y., Lei, A. & Gabrilovich, D. Neutrophils and PMN-MDSC: their biological role and interaction with stromal cells. Semin. Immunol. 35, 19–28 (2018).
Blatner, N. R. et al. Expression of RORγt marks a pathogenic regulatory T cell subset in human colon cancer. Sci. Transl. Med. 4, 164ra159 (2012).
Miragaia, R. J. et al. Single-cell transcriptomics of regulatory T cells reveals trajectories of tissue adaptation. Immunity 50, 493–504 (2019).
Blatner, N. R. et al. In colorectal cancer mast cells contribute to systemic regulatory T cell dysfunction. Proc. Natl Acad. Sci. USA 107, 6430–6435 (2010).
Gounaris, E. et al. T regulatory cells shift from a protective anti-inflammatory to a cancer-promoting proinflammatory phenotype in polyposis. Cancer Res. 69, 5490–5497 (2009).
Keerthivasan, S. et al. β-Catenin promotes colitis and colon cancer through imprinting of proinflammatory properties in T cells. Sci. Transl. Med. 6, 225ra228 (2014).
Quandt, J. et al. Wnt–β-catenin activation epigenetically reprograms Treg cells in inflammatory bowel disease and dysplastic progression. Nat. Immunol. 22, 471–484 (2021).
Sumida, T. et al. Activated β-catenin in Foxp3+ regulatory T cells links inflammatory environments to autoimmunity. Nat. Immunol. 19, 1391–1402 (2018).
Mosimann, C., Hausmann, G. & Basler, K. β-catenin hits chromatin: regulation of Wnt target gene activation. Nat. Rev. Mol. Cell Biol. 10, 276–286 (2009).
Barra, M. M., Richards, D. M., Hofer, A. C., Delacher, M. & Feuerer, M. Premature expression of Foxp3 in double-negative thymocytes. PLoS ONE 10, e0127038 (2015).
Barra, M. M. et al. Transcription factor 7 limits regulatory T cell generation in the thymus. J. Immunol. 195, 3058–3070 (2015).
van Loosdregt, J. et al. Canonical Wnt signaling negatively modulates regulatory T cell function. Immunity 39, 298–310 (2013).
Xing, S. et al. Tcf1 and Lef1 are required for the immunosuppressive function of regulatory T cells. J. Exp. Med. 216, 847–866 (2019).
Mielke, L. A. et al. TCF-1 limits the formation of Tc17 cells via repression of the MAF–RORγt axis. J. Exp. Med. 216, 1682–1699 (2019).
Emmanuel, A. O. et al. TCF-1 and HEB cooperate to establish the epigenetic and transcription profiles of CD4+CD8+ thymocytes. Nat. Immunol. 19, 1366–1378 (2018).
Rubtsov, Y. P. et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558 (2008).
Chapman, N. M. & Chi, H. mTOR links environmental signals to T cell fate decisions. Front Immunol. 5, 686 (2014).
Neumann, C. et al. c-Maf-dependent Treg cell control of intestinal TH17 cells and IgA establishes host–microbiota homeostasis. Nat. Immunol. 20, 471–481 (2019).
Kim, H. J. et al. Stable inhibitory activity of regulatory T cells requires the transcription factor Helios. Science 350, 334–339 (2015).
Thornton, A. M. et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J. Immunol. 184, 3433–3441 (2010).
Fassett, M. S., Jiang, W., D’Alise, A. M., Mathis, D. & Benoist, C. Nuclear receptor Nr4a1 modulates both regulatory T cell differentiation and clonal deletion. Proc. Natl Acad. Sci. USA 109, 3891–3896 (2012).
Kovalovsky, D. et al. β-Catenin/Tcf determines the outcome of thymic selection in response to αβTCR signaling. J. Immunol. 183, 3873–3884 (2009).
Pabbisetty, S. K. et al. Peripheral tolerance can be modified by altering KLF2-regulated Treg migration. Proc. Natl Acad. Sci. USA 113, E4662–E4670 (2016).
Beischlag, T. V. et al. Recruitment of the NCoA/SRC-1/p160 family of transcriptional coactivators by the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator complex. Mol. Cell. Biol. 22, 4319–4333 (2002).
Zemmour, D. et al. Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat. Immunol. 19, 291–301 (2018).
Balderhaar, H. J. et al. The CORVET complex promotes tethering and fusion of Rab5/Vps21-positive membranes. Proc. Natl Acad. Sci. USA 110, 3823–3828 (2013).
Kolinski, T., Marek-Trzonkowska, N., Trzonkowski, P. & Siebert, J. Heat-shock proteins (HSPs) in the homeostasis of regulatory T cells. Cent. Eur. J. Immunol. 41, 317–323 (2016).
Joller, N. et al. Treg cells expressing the coinhibitory molecule TIGIT selectively inhibit proinflammatory TH1 and TH17 cell responses. Immunity 40, 569–581 (2014).
Qiu, X. B., Shao, Y. M., Miao, S. & Wang, L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 63, 2560–2570 (2006).
Weis, A. M., Soto, R. & Round, J. L. Commensal regulation of T cell survival through Erdr1. Gut Microbes 9, 458–464 (2018).
Miyagawa, I. et al. Induction of regulatory T cells and its regulation with insulin-like growth factor/insulin-like growth factor binding protein-4 by human mesenchymal stem cells. J. Immunol. 199, 1616–1625 (2017).
Bettelli, E. et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238 (2006).
La Manno, G. et al. RNA velocity of single cells. Nature 560, 494–498 (2018).
Chen, Z. et al. Selective regulatory function of Socs3 in the formation of IL-17-secreting T cells. Proc. Natl Acad. Sci. USA 103, 8137–8142 (2006).
Meixner, A., Karreth, F., Kenner, L. & Wagner, E. F. JunD regulates lymphocyte proliferation and T helper cell cytokine expression. EMBO J. 23, 1325–1335 (2004).
Woo, S. R. et al. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T cell function to promote tumoral immune escape. Cancer Res. 72, 917–927 (2012).
Chen, M. L. et al. Regulatory T cells suppress tumor-specific CD8+ T cell cytotoxicity through TGF-β signals in vivo. Proc. Natl Acad. Sci. USA 102, 419–424 (2005).
Fahlen, L. et al. T cells that cannot respond to TGF-β escape control by CD4+CD25+ regulatory T cells. J. Exp. Med. 201, 737–746 (2005).
Mempel, T. R. et al. Regulatory T cells reversibly suppress cytotoxic T cell function independent of effector differentiation. Immunity 25, 129–141 (2006).
Pavelko, K. D. et al. Theiler’s murine encephalomyelitis virus as a vaccine candidate for immunotherapy. PLoS ONE 6, e20217 (2011).
Esplugues, E. et al. Control of TH17 cells occurs in the small intestine. Nature 475, 514–518 (2011).
Gounaris, E. et al. Live imaging of cysteine–cathepsin activity reveals dynamics of focal inflammation, angiogenesis and polyp growth. PLoS ONE 3, e2916 (2008).
Zhang, Y. et al. Deep single-cell RNA-sequencing data of individual T cells from treatment-naive colorectal cancer patients. Sci. Data 6, 131 (2019).
Blatner, N. R., Gounari, F. & Khazaie, K. The two faces of regulatory T cells in cancer. Oncoimmunology 2, e23852 (2013).
Bos, P. D. & Rudensky, A. Y. Treg cells in cancer: a case of multiple personality disorder. Sci. Transl. Med. 4, 164fs144 (2012).
Kwon, H. K., Chen, H. M., Mathis, D. & Benoist, C. Different molecular complexes that mediate transcriptional induction and repression by Foxp3. Nat. Immunol. 18, 1238–1248 (2017).
van der Veeken, J. et al. The transcription factor Foxp3 shapes regulatory T cell identity by tuning the activity of trans-acting intermediaries. Immunity 53, 971–984 (2020).
Pavelko, K. D., Bell, M. P., Harrington, S. M. & Dong, H. B7-H1 influences the accumulation of virus-specific tissue resident memory T cells in the central nervous system. Front. Immunol. 8, 1532 (2017).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016).
Acknowledgements
This work was supported by NIH grants R01 AI 108682 (to F.G. and K.K.), RO1 AI 147652 (to F.G.) and R35GM138283 (to M.K.), and a Praespero Innovation Award Alberta, Canada (to F.G. and K.K.). N. Carapanceanu and V. Carapanceanu are thankfully acknowledged for excellent technical support. L. Pennell (BioLegend) is gratefully acknowledged for advice with scRNA-seq techniques. V. Simon (Mayo Clinic) is gratefully acknowledged for assistance with scRNA-seq. A. V. Lucs (Eli Lilly) is thankfully acknowledged for providing the TGF-βRI inhibitor LY3200882 and for scientific advice.
Author information
Authors and Affiliations
Contributions
A.O. planned and performed experiments; acquired, analyzed and interpreted data; and helped with writing of the manuscript. K.D.P., J.Q., A.S. and M.P.S. performed experiments, acquired and analyzed data, and prepared figures. B.Y. and Y.L. performed scRNA-seq analysis and prepared figures, helped with the interpretation of data and writing of the manuscript. M.K., F.G. and K.K. analyzed and interpreted data, prepared figures and wrote the manuscript. K.K. conceived the project and designed and oversaw experiments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Immunology thanks Federica Facciotti and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. L. A. Dempsey was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 TCF-1 deficiency selectively reprograms Treg-cells without compromising their core signature.
Treg-cells were isolated from the mesenteric lymph nodes of Foxp3Cre and Tcf7fl/flFoxp3Cre mice. (a) Representative FACS histograms of MLN purified cells from Foxp3Cre Tcf7fl/fl and control Foxp3Cre showing selective loss of TCF-1 from Treg-cells in Foxp3Cre Tcf7fl/fl mice. (b and c) Histogram plots showing the cumulative data of the same. (b: n = 4; p < 0.0001 & c: n = 5) Data are representative of two independent experiments and n represents biologically independent replicate mice; means ± SEM; two-sided, unpaired t-test. (d) GSEA plot comparing the enrichment of genes expressed more strongly in Foxp3Cre versus Foxp3CreTcf7fl/fl Treg-cells.
Extended Data Fig. 2 Representative FACS plots of lymphocyte surface markers expressed by Treg-cells.
Treg-cells were isolated from the mesenteric lymph nodes of Foxp3Cre and Tcf7fl/flFoxp3Cre mice. See cumulative data presented in Fig. 2. (a-c) CD4+ cells were pre-gated and frequency of CD69+, ICOS+, and PD1+ cells among CD4+FOXP3− Tcon or CD4+FOXP3+ Treg-cells was measured, as indicated. (d) CD4+ cells were pre-gated and frequency of CD44+CD62L− cells among CD4+FOXP3− Tcon cells was measured. (e) CD4+ cells were pre-gated and frequency of CD4+FOXP3+ Treg-cells was measured. (f) CD4+ cells were pre-gated and frequency of FOXP3+CD25+ Treg-cells was measured. (g) CD4+FOXP3+ Treg-cells were pre-gated and frequency of RORγT+HELIOS− or RORγT+HELIOS+ was measured. (h) CD4+FOXP3+ Treg-cells and frequency of CD44+CD62L− cells among Treg cells was measured. Numbers inside quadrants indicate percent cells in the respective quadrants.
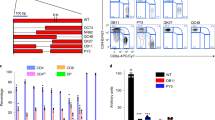
Extended Data Fig. 3 Treg purification.
Treg-cells were isolated from the mesenteric lymph nodes of Foxp3Cre and Tcf7fl/flFoxp3Cre mice. (a) Schematic representation of purification of Treg-cells, and FACS analysis showing over 90% purity. (b) Expression changes of the Tcf7 transcripts between TCF-1-deficient and TCF-1-sufficient Treg-cells. The color intensity is proportional to the average gene expression across cells in the indicated Treg cluster. The size of circles is proportional to percentage of cells expressing indicated genes.
Extended Data Fig. 4 Single-cell RNAseq reveals distinct Treg populations.
mRNA expression of select indicated genes projected on the UMAP. Note varied expression of Klf2 but broad and uniform expression of Izumo1r by Treg clusters, high expression of Mif, Vps8, and Ifit1 in the respective Mif (cluster 3), Vps8 (cluster 8), Ifn (cluster 9). Expression of Ccl5 is prominent in the Cd63 (cluster 7), which is likely not Treg-cells.
Extended Data Fig. 5 TCF-1-deficient and sufficient Treg-cells show distinct effector functions.
Treg-cells were isolated from the mesenteric lymph nodes of Foxp3Cre and Tcf7fl/flFoxp3Cre mice. (a) mRNA expression of Maf projected on the UMAP, comparing Treg-cells derived from Foxp3Cre to Tcf7fl/fl Foxp3Cre mice. (b) Violin plots showing expression of Maf in individual Treg clusters. (c) GSEA of MAF downregulated genes and TH17 pathway defined by Stubbington. (d) Kegg IL17 signaling pathway projected on UMAP, comparing TCF-1-sufficient and TCF-1-deficient Treg-cells (e) GSEA analysis for the Kegg IL17 signaling pathway comparing transcriptomes of TCF-1-sufficient and TCF-1-deficient Treg-cells across all cell types. Normalized enrichment scores (NES) are color coded. -log10 (FDR) values are proportional to the circle size. FDR > 15% are masked with gray color. (fgh) mRNA expression of Ccr9, Erdr1 and Igfbp4 projected on the UMAP, comparing TCF-1-sufficient and TCF-1-deficient Klf2– cells for the Kegg IL17 pathway.
Extended Data Fig. 6 Treg-cells are activated and polarized during polyposis.
Treg_cells were isolated from the mesenteric lymph nodes of WT and APCΔ486 mice. (a) UMAP projection (left panel) and fraction of cells in each cell type (stack bars; right panel) for APCΔ486 and control B6 Treg-cells. Data are from two replicates. (b) Dot plot showing the expression of Tcf7 across all cell types in ApcΔ486 and control B6 Treg-cells. Color and size of the dots are proportional to the expression level and percent of cells expressing Tcf7 in each indicated cluster. (c) Expression of Socs3, Jund, Lag3 and Maf between APCΔ486 and B6 cells projected on the UMAP. See TableS4 for the full list. The fold change in percent of cells expressing the indicated gene in each cell type is proportional to the circle size. Adjusted-p-values > 0.01 are masked with gray color. (d) Expression changes of the most differentially expressed genes between APCΔ486 and control B6 Treg-cells. See TableS4 for the full list. The fold change in expression intensities is color-coded. (e) RNA velocity vectors overlaid on UMAP for B6 (left) and APCΔ486 (right) Treg-cells.
Supplementary information
Source data
Source Data Fig. 2
Numerical FACS values.
Source Data Fig. 5
Numerical FACS values.
Source Data Fig. 6
Numerical FACS values.
Source Data Fig. 7
Numerical FACS values.
Source Data Extended Data Fig. 1
Numerical FACS values.
Rights and permissions
About this article
Cite this article
Osman, A., Yan, B., Li, Y. et al. TCF-1 controls Treg cell functions that regulate inflammation, CD8+ T cell cytotoxicity and severity of colon cancer. Nat Immunol 22, 1152–1162 (2021). https://doi.org/10.1038/s41590-021-00987-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-021-00987-1
This article is cited by
-
The dynamic shifts of IL-10-producing Th17 and IL-17-producing Treg in health and disease: a crosstalk between ancient "Yin-Yang" theory and modern immunology
Cell Communication and Signaling (2024)
-
The regulation and differentiation of regulatory T cells and their dysfunction in autoimmune diseases
Nature Reviews Immunology (2024)
-
Pertussis toxin-induced inhibition of Wnt/β-catenin signaling in dendritic cells promotes an autoimmune response in experimental autoimmune uveitis
Journal of Neuroinflammation (2023)
-
Regulatory T cells in the face of the intestinal microbiota
Nature Reviews Immunology (2023)
-
Ubiquitinomics revealed disease- and stage-specific patterns relevant for the 3PM approach in human sigmoid colon cancers
EPMA Journal (2023)