Abstract
Fibroblasts are one of the most common but also neglected types of stromal cells, the heterogeneity of which underlies the specific function of tissue microenvironments in development and regeneration. In the thymus, autoreactive T cells are thought to be negatively selected by reference to the self-antigens expressed in medullary epithelial cells, but the contribution of other stromal cells to tolerance induction has been poorly examined. In the present study, we report a PDGFR+ gp38+ DPP4− thymic fibroblast subset that is required for T cell tolerance induction. The deletion of the lymphotoxin β-receptor in thymic fibroblasts caused an autoimmune phenotype with decreased expression of tissue-restricted and fibroblast-specific antigens, offering insight into the long-sought target of lymphotoxin signaling in the context of the regulation of autoimmunity. Thus, thymic medullary fibroblasts play an essential role in the establishment of central tolerance by producing a diverse array of self-antigens.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
RNA-seq data are available in the GEO database (www.ncbi.nlm.nih.gov/geo) with accession no. GSE147357. The full code for single-cell RNA-seq analysis is available in GitHub (https://github.com/nittatakeshi/scRNAseq.git). TCR sequence data have been deposited in the Sequence Read Archives (www.ncbi.nlm.nih.gov/sra) with BioProject ID no. PRJNA641239. All additional data that support the findings of the present study are available from the corresponding author upon request. Source data are provided with this paper.
References
Abramson, J. & Anderson, G. Thymic epithelial cells. Annu. Rev. Immunol. 35, 85–118 (2017).
Takahama, Y., Ohigashi, I., Baik, S. & Anderson, G. Generation of diversity in thymic epithelial cells. Nat. Rev. Immunol. 17, 295–305 (2017).
Bornstein, C. et al. Single-cell mapping of the thymic stroma identifies IL-25-producing tuft epithelial cells. Nature 559, 622–626 (2018).
Miller, C. N. et al. Thymic tuft cells promote an IL-4-enriched medulla and shape thymocyte development. Nature 559, 627–631 (2018).
Gray, D. H. et al. A unique thymic fibroblast population revealed by the monoclonal antibody MTS-15. J. Immunol. 178, 4956–4965 (2007).
Muller, S. M. et al. Neural crest origin of perivascular mesenchyme in the adult thymus. J. Immunol. 180, 5344–5351 (2008).
Foster, K. et al. Contribution of neural crest-derived cells in the embryonic and adult thymus. J. Immunol. 180, 3183–3189 (2008).
Fuertbauer, E. et al. Thymic medullar conduits-associated podoplanin promotes natural regulatory T cells. Immunol. Lett. 154, 31–41 (2013).
Lynch, M. D. & Watt, F. M. Fibroblast heterogeneity: implications for human disease. J. Clin. Invest. 128, 26–35 (2018).
Guerrero-Juarez, C. F. et al. Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nat. Commun. 10, 650 (2019).
Mizoguchi, F. et al. Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat. Commun. 9, 789 (2018).
Wohlfahrt, T. et al. PU.1 controls fibroblast polarization and tissue fibrosis. Nature 566, 344–349 (2019).
Perez-Shibayama, C., Gil-Cruz, C. & Ludewig, B. Fibroblastic reticular cells at the nexus of innate and adaptive immune responses. Immunol. Rev. 289, 31–41 (2019).
Boehm, T., Scheu, S., Pfeffer, K. & Bleul, C. C. Thymic medullary epithelial cell differentiation, thymocyte emigration, and the control of autoimmunity require lympho-epithelial cross talk via LTβR. J. Exp. Med. 198, 757–769 (2003).
Chin, R. K. et al. Lymphotoxin pathway-directed, autoimmune regulator-independent central tolerance to arthritogenic collagen. J. Immunol. 177, 290–297 (2006).
Martins, V. C., Boehm, T. & Bleul, C. C. LTβR signaling does not regulate Aire-dependent transcripts in medullary thymic epithelial cells. J. Immunol. 181, 400–407 (2008).
Balciunaite, G. et al. Wnt glycoproteins regulate the expression of FoxN1, the gene defective in nude mice. Nat. Immunol. 3, 1102–1108 (2002).
Hikosaka, Y. et al. The cytokine RANKL produced by positively selected thymocytes fosters medullary thymic epithelial cells that express autoimmune regulator. Immunity 29, 438–450 (2008).
Seach, N. et al. The lymphotoxin pathway regulates Aire-independent expression of ectopic genes and chemokines in thymic stromal cells. J. Immunol. 180, 5384–5392 (2008).
White, A. J. et al. Lymphotoxin signals from positively selected thymocytes regulate the terminal differentiation of medullary thymic epithelial cells. J. Immunol. 185, 4769–4776 (2010).
Wu, W. et al. Epithelial LTβR signaling controls the population size of the progenitors of medullary thymic epithelial cells in neonatal mice. Sci. Rep. 7, 44481 (2017).
Cosway, E. J. et al. Redefining thymus medulla specialization for central tolerance. J. Exp. Med. 214, 3183–3195 (2017).
Jenkinson, W. E., Rossi, S. W., Parnell, S. M., Jenkinson, E. J. & Anderson, G. PDGFRα-expressing mesenchyme regulates thymus growth and the availability of intrathymic niches. Blood 109, 954–960 (2007).
Suniara, R. K., Jenkinson, E. J. & Owen, J. J. An essential role for thymic mesenchyme in early T cell development. J. Exp. Med. 191, 1051–1056 (2000).
Itoi, M., Tsukamoto, N., Yoshida, H. & Amagai, T. Mesenchymal cells are required for functional development of thymic epithelial cells. Int. Immunol. 19, 953–964 (2007).
Sitnik, K. M. et al. Mesenchymal cells regulate retinoic acid receptor-dependent cortical thymic epithelial cell homeostasis. J. Immunol. 188, 4801–4809 (2012).
Van Vliet, E., Melis, M. & Van Ewijk, W. Monoclonal antibodies to stromal cell types of the mouse thymus. Eur. J. Immunol. 14, 524–529 (1984).
Odaka, C. Localization of mesenchymal cells in adult mouse thymus: their abnormal distribution in mice with disorganization of thymic medullary epithelium. J. Histochem. Cytochem. 57, 373–382 (2009).
Sun, L. et al. FSP1+ fibroblast subpopulation is essential for the maintenance and regeneration of medullary thymic epithelial cells. Sci. Rep. 5, 14871 (2015).
Shores, E. W., Van Ewijk, W. & Singer, A. Disorganization and restoration of thymic medullary epithelial cells in T cell receptor-negative scid mice: evidence that receptor-bearing lymphocytes influence maturation of the thymic microenvironment. Eur. J. Immunol. 21, 1657–1661 (1991).
van Ewijk, W., Shores, E. W. & Singer, A. Crosstalk in the mouse thymus. Immunol. Today 15, 214–217 (1994).
Akiyama, T. et al. The tumor necrosis factor family receptors RANK and CD40 cooperatively establish the thymic medullary microenvironment and self-tolerance. Immunity 29, 423–437 (2008).
Fletcher, A. L. et al. Lymph node fibroblastic reticular cells directly present peripheral tissue antigen under steady-state and inflammatory conditions. J. Exp. Med. 207, 689–697 (2010).
Chen, Y. et al. CXCR5+PD-1+ follicular helper CD8 T cells control B cell tolerance. Nat. Commun. 10, 4415 (2019).
Perry, J. S. A. et al. Distinct contributions of Aire and antigen-presenting-cell subsets to the generation of self-tolerance in the thymus. Immunity 41, 414–426 (2014).
Dubrot, J. et al. Lymph node stromal cells acquire peptide–MHCII complexes from dendritic cells and induce antigen-specific CD4+ T cell tolerance. J. Exp. Med. 211, 1153–1166 (2014).
Kedl, R. M. et al. Migratory dendritic cells acquire and present lymphatic endothelial cell-archived antigens during lymph node contraction. Nat. Commun. 8, 2034 (2017).
Klein, L., Klein, T., Ruther, U. & Kyewski, B. CD4 T cell tolerance to human C-reactive protein, an inducible serum protein, is mediated by medullary thymic epithelium. J. Exp. Med. 188, 5–16 (1998).
Derbinski, J., Schulte, A., Kyewski, B. & Klein, L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat. Immunol. 2, 1032–1039 (2001).
Derbinski, J. et al. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J. Exp. Med. 202, 33–45 (2005).
St-Pierre, C., Trofimov, A., Brochu, S., Lemieux, S. & Perreault, C. Differential features of AIRE-induced and AIRE-independent promiscuous gene expression in thymic epithelial cells. J. Immunol. 195, 498–506 (2015).
Sosic, D., Richardson, J. A., Yu, K., Ornitz, D. M. & Olson, E. N. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-κB activity. Cell 112, 169–180 (2003).
Gordon, J. et al. Specific expression of lacZ and cre recombinase in fetal thymic epithelial cells by multiplex gene targeting at the Foxn1 locus. BMC Dev. Biol. 7, 69 (2007).
Kawabe, T. et al. The immune responses in CD40-deficient mice: impaired immunoglobulin class switching and germinal center formation. Immunity 1, 167–178 (1994).
Mombaerts, P. et al. Mutations in T-cell antigen receptor genes α and β block thymocyte development at different stages. Nature 360, 225–231 (1992).
Mombaerts, P. et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68, 869–877 (1992).
Kawamoto, S. et al. A novel reporter mouse strain that expresses enhanced green fluorescent protein upon Cre-mediated recombination. FEBS Lett. 470, 263–268 (2000).
Hanada, R. et al. Central control of fever and female body temperature by RANKL/RANK. Nature 462, 505–509 (2009).
Mouri, Y. et al. Lymphotoxin signal promotes thymic organogenesis by eliciting RANK expression in the embryonic thymic stroma. J. Immunol. 186, 5047–5057 (2011).
Shinkura, R. et al. Alymphoplasia is caused by a point mutation in the mouse gene encoding Nf-κb-inducing kinase. Nat. Genet. 22, 74–77 (1999).
Nitta, T. et al. The thymic cortical epithelium determines the TCR repertoire of IL-17-producing γδ T cells. EMBO Rep. 16, 638–653 (2015).
Seach, N., Wong, K., Hammett, M., Boyd, R. L. & Chidgey, A. P. Purified enzymes improve isolation and characterization of the adult thymic epithelium. J. Immunol. Methods 385, 23–34 (2012).
Nitta, T., Ohigashi, I. & Takahama, Y. The development of T lymphocytes in fetal thymus organ culture. Methods Mol. Biol. 946, 85–102 (2013).
Nitta, T. et al. Human thymoproteasome variations influence CD8 T cell selection. Sci. Immunol. 2, eaan5165 (2017).
Schug, J. et al. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 6, R33 (2005).
Tomofuji, Y. et al. Chd4 choreographs self-antigen expression for central immune tolerance. Nat. Immunol. 21, 892–901 (2020).
Acknowledgements
We thank K. Kubo, Y. Nakayama, R. Yanobu-Takanashi, K. Kaneki, Y. Yoshitomi, R. Sakuma, S. Igaue, K. Ikeda, T. Iguchi, N. C-N. Huynh, M. Tsukasaki and A. Ota for technical assistance and all laboratory members for insightful discussions. Special thanks to N. Akiyama and T. Akiyama for technical support in thymus transplant experiments. This work was supported by the Japan Society for Promotion of Science (grant nos. KAKENHI 15H05703 to H.T. and 16H05202, 17H05788, 19H03485 and 19H04802 to T.N.), the CREST program of the Japan Agency for Medical Research and Development (grant no. 20gm1210008 to H.T.), the Tokyo Society of Medical Science (to T.N.) and the National Center for Global Health and Medicine (grant nos. 26-105 and 29-1001 to T.O.).
Author information
Authors and Affiliations
Contributions
T.N. and H.T. conceived the ideas and designed the experiments. T.N., M.T., S.N., R.M. and E.C.S. performed the experiments. E.C.S. and Y.T. performed bioinformatic analysis. K.N., T.O. and J.M.P. generated genetically modified mice. S.S. provided advice on project design and data interpretation. T.N. and H.T. wrote the manuscript. H.T. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interest.
Additional information
Peer review information L. A. Dempsey was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Enzymatic fractionation of thymic cells.
a, Thymic cells were isolated by gradual enzymatic digestion as shown in Fig. 1d. Cells from each fraction were measured by flow cytometry analysis (mean ± SEM, three independent experiments). To examine thymocyte subpopulations, cells were stained for CD3, CD25, TCRβ, and CD24 instead of CD4 and CD8, because these two proteins are sensitive to Liberase digestion. Numbers of DN2-3 thymocytes (CD3- CD25+, n=3), DP thymocytes (CD3-/lo TCRβ-/lo, n=3), SP thymocytes (CD3+ TCRβ+ CD24-, n=3), γδT cells (CD3+ TCRβ- CD24-, n=3), regulatory T cells (Treg, CD3+ CD25+, n=3), dendritic cells (DC, CD11c+ MHC-II+, n=4), B cells (CD19+ MHC-II+, n=3), and endothelial cells (EC, CD45- EpCAM- CD31+, n=4) in each fraction are shown. b, Whole thymic cells were prepared by Liberase digestion and analyzed by flow cytometry. Histograms indicate surface expression of H2-Kb (MHC-I), I-Ab (MHC-II), and CD80 on gated capFbs, mFbs, and TECs. The numbers indicate the mean fluorescence intensity (MFI). Data shown are representative of three independent experiments.
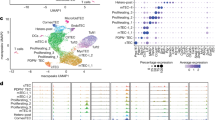
Extended Data Fig. 2 Transcriptome analysis of fibroblast subsets.
a, Scatter plot of bulk RNA-seq data comparing mRNA expression levels between capFb and mFb. Selected capFb- or mFb-specific genes are shown. b, Heat map of selected chemokine genes and Wnt ligand genes expressed in capFbs and mFbs. (c and d) Single-cell RNA-seq data of thymic stromal cells (GSE103967, Experiment ID thymus_stroma_WT) 3 were used for quality control and cell-cycle correction, followed by t-SNE analysis. c, Two-dimensional representation of cells via t-SNE. Each dot represents one cell. d, Projection of representative genes on the two-dimensional graphs. Clusters 2, 7, and 10 are thymic fibroblasts characterized by the expression of Pdgfra, Pdgfrb, and Cd34. Clusters 7 and 10 represent capFbs expressing Dpp4, Pi16, and Mfap5, while cluster 2 represents mFbs expressing Col6a5, Col6a6, Mmp9, Ltbp1, Enpp2, and Hmgcs2. Clusters 1, 3, 6, and 9 represent TECs. Cluster 4 represents endothelial cells (ECs) expressing Pecam1 (CD31). These data also include contaminating lymphocytes (cluster 5) defined by high expression of lymphocyte-specific genes such as Cd3e. e, Isolation of LN FRCs. sLN (pooled axillary and inguinal LNs) or mLN isolated from 8-week-old WT mice were dissociated with 0.01% Liberase TM and 0.01% DNase I. CD45+ cells and Ter119+ cells were magnetically depleted. Representative flow cytometry profiles of pre-depletion, post-depletion, and post-depletion CD45- stromal cells are shown. FRCs (gp38+CD31-, indicated with red boxes) were isolated and subjected to RNA-seq analysis (Fig. 3d–f).
Extended Data Fig. 3 Thymic stromal cells in mice lacking SP thymocytes or mTECs.
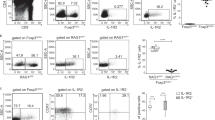
a, Flow cytometry analysis of thymic fibroblasts from embryonic day (E) 13.5 (n=2), E14.5 (n=3), E15.5 (n=5) or E17.5 (n=5) of WT mice. CD45-EpCAM-CD31-PDGFR+ cells were analyzed for gp38 and DPP4 expression. Data represent two independent experiments per group. b, Thymus sections from E15.5 WT mice were stained for gp38, DPP4, and K14. Data are representative of two independent experiments. c, Thymus sections from 0-day-old WT mice were stained for gp38, DPP4, and K14. DPP4 expression was strongly detected in gp38-expressing capFbs and weakly detected in thymocytes. Data are representative of two independent experiments. (d and e) Five to 8-week-old WT, Tcra-/-, or Foxn1-Cre Tnfrsf11aflox/flox Cd40-/- mice were analyzed. d, Thymus sections were stained for gp38, CD205, K14, CD4, and CD8. First and second lines are identical sections. Data are representative of two independent experiments. e, Flow cytometry profiles of gated TECs (CD45- EpCAM+) for Ly51 and UEA1 and gated mTECs (CD45- EpCAM+ Ly51- UEA1+) for MHC-II and CD80. Graphs show numbers of indicated TEC subsets (cells per thymus lobe). WT (n=9), Tcra-/- (n=7), or Foxn1-Cre Tnfrsf11aflox/flox Cd40-/- (n=4) mice were analyzed. Significance was determined by one-way ANOVA with Holm-Sidak’s multiple comparison test.
Extended Data Fig. 4 Conditional gene-targeting of Ltbr.
a, capFbs (n=4), mFbs (n=4), cTECs (n=3), mTECs (n=3), endothelial cells (EC) (n=4), pericytes (PC) (n=4), and CD45+ lymphocytes (Lym) (n=3) were sorted from 5-week-old WT mice and analyzed for Ltbr mRNA expression by RT-qPCR. Ltbr mRNA expression was normalized to Gapdh mRNA, and that in total thymus was arbitrarily set to 1 (mean ± SEM). b, Scheme of the targeting strategy. Two sgRNAs and two loxP-containing oligonucleotides were designed to insert the loxP sequences into the introns 2 and 5 of Ltbr. c, The target sequences of sgRNAs and oligonucleotides for 5’ and 3’ loxP sites. The loxP sequence was underlined. d, Flow cytometry analysis of LTβR expression in thymic stromal cells from Ltbrflox, LtbrΔFb, and Ltbr-/- mice (top). MFI of LTβR expression (mean ± SEM) (bottom). Significance was determined by one-way ANOVA with Holm-Sidak’s multiple comparison test. e, Representative photographs of the indicated lymph nodes in Ltbrflox or LtbrΔFb mice. Black or red arrows indicate presence or absence of the lymph nodes, respectively. f, The presence of the indicated lymph nodes was plotted as percent present/absent. For axillary and inguinal lymph nodes, both sides (left and right) were examined.
Extended Data Fig. 5 Autoantibody production in fibroblast-specific LTβR-deficient mice.
a, Autoantibody production in LTβR-deficient mice. Tissue sections from Rag1-/- mice were stained with serum from 4-8-month-old Ltbrflox, Ltbr-/-, LtbrΔFb, or LtbrΔTEC mice. For lung, two representative images for each mouse strains, because the staining patterns were different among individual mice. Some mice exhibited nuclear staining, while others showed cytoplasmic or extracellular staining patterns. Data shown represent three independent experiments using Ltbrflox (n=6), Ltbr-/- (n=6), LtbrΔFb (n=11), and LtbrΔTEC (n=8) mice for lung, pancreas, and salivary gland, or Ltbrflox (n=9), Ltbr-/- (n=10), LtbrΔFb (n=8), and LtbrΔTEC (n=8) mice for liver. Autoantibody staining intensity was quantified based on an arbitrary scale of 0-4, as shown in Fig. 5b. b, Higher magnification of framed area in (a). c, Kidney sections from the indicated mice were stained with anti-IgM or anti-IgG antibodies. Ltbr-/- and LtbrΔFb mice exhibited markedly elevated IgM deposition in the glomeruli. Data shown represent two independent experiments using Ltbrflox (n=2), Ltbr-/- (n=3), and LtbrΔFb (n=3) mice.
Extended Data Fig. 6 LTβR-dependent genes in mFbs include TRAs.
Heat map shows the relative expression levels of selected LTβR-dependent genes in mFbs and mTEChi cells from Ltbrflox or LtbrΔFb mice. The gene expression in cDC1 (CD11chi PDCA1lo Sirpα-), cDC2 (CD11chi PDCA1lo Sirpα+), and pDC (CD11clo PDCA1hi) are also shown. The tissue-specific expression of each gene was based on the TiGER database and entropy score.
Extended Data Fig. 7 Fibroblasts in the grafted thymus.
dGuo-treated fetal thymuses from Ltbrflox mice (n=4) or LtbrΔFb mice (n=7) were transplanted in kidney capsule of athymic Foxn1-/- mice, as shown in Fig. 7b. a, Representative flow cytometry profiles of gated fibroblasts (CD45-EpCAM-CD31-PDGFR+) for gp38 and DPP4 expression in grafted thymuses. b, LTβR expression levels in gated gp38+DPP4+ cells (left) and gp38+DPP4- cells (center) from grafted thymuses. gp38 expression levels in gated gp38+DPP4- cells from Ltbrflox grafts and gp38+DPP4-LTβR- cells from LtbrΔFb grafts (right). Significance was determined by unpaired two-tailed t-test. As stated in the text, the autoimmune phenotype was less pronounced in the LtbrΔFb thymus-grafted mice compared to that in LtbrΔFb mice, possibly because of the repopulation by host-derived fibroblasts. It was previously reported that, when transplanted in kidney capsule, the grafted thymus can be invaded and repopulated by host-derived mesenchymal cells (likely kidney fibroblasts)23. The data indicate that the expression levels of LTβR in gp38+DPP4+ cells were comparable between LtbrΔFb and Ltbrflox grafts, indicating that these cells are mainly host-derived fibroblasts (b, left panel). In contrast, gp38+DPP4- cells in LtbrΔFb grafts contained LTβR-non-expressing cells (51±4%) as well as LTβR-expressing cells (b, center panel), indicating that these cells are a mixture of donor-derived (LTβR-deficient) mFbs and host-derived (WT) fibroblasts. The gp38+DPP4-LTβR- cells in LtbrΔFb grafts showed significantly lower levels of gp38 expression compared with gp38+DPP4- cells in Ltbrflox grafts (b, right panel), representing a phenotype that recapitulates that of mFbs in LtbrΔFb mice (Fig. 6a).
Extended Data Fig. 8 mTECs in fibroblast-specific LTβR-deficient mice.
(a and b) Thymus sections from 5-8-week-old Ltbrflox, Ltbr-/-, LtbrΔFb, LtbrΔTEC mice were stained with hematoxylin and eosin (a) or for indicated markers (b). Data represent two independent experiments. c, Thymus sections from 8-week-old Ltbrflox or LtbrΔFb mice were stained with anti-CD205 (cTEC marker) and UEA1 (mTEC marker). The staining intensity of UEA1 per unit area of the medullary region (indicated by yellow lines) was calculated. Three representative images from three individual mice per group were examined. Significance was determined by unpaired two-tailed t-test. d, Thymus sections from 8-week-old Ltbrflox or LtbrΔFb mice were stained with anti-CD205 (cTEC marker), anti-K14 (mTEC marker), and anti-Aire. The number of Aire+ cells per unit area of the medullary region (indicated by yellow lines) was calculated. Three representative images from three individual mice per group were examined. Significance was determined by unpaired two-tailed t-test. e–g, mTEChi and mTEClo cells isolated from Ltbrflox, LtbrΔFb or LtbrΔTEC mice (n=2 per each group) were analyzed by RNA-seq. e, Principle component analysis of RNA-seq data. This data suggests that there is no marked difference in gene expression in mTEChi cells among the three groups, while mTEClo cells of LtbrΔTEC mice show a gene expression profile different from those of Ltbrflox or LtbrΔFb mice. f, Scatter plots for mRNA expression levels. The numbers of up-regulated or down-regulated genes (fold change ≥ 5) are shown. g, Heat map of selected genes expressed in mTECs.
Extended Data Fig. 9 Thymic dendritic cells in LTβR-deficient mice.
a, Thymus sections from 8-week-old Ltbrflox, Ltbr-/-, LtbrΔFb, or LtbrΔTEC mice were stained for K14 (mTECs) and CD11c (DCs). Data represent two independent experiments. b, Representative flow cytometry profiles for CD11c and PDCA1 expression of whole thymic cells. Frequencies of CD11chi PDCA1lo cells (cDCs) and CD11clo PDCA1hi cells (pDCs) are indicated. c, Numbers of total thymic DCs, cDCs, and pDCs (cells per mouse) in Ltbrflox (n=10), Ltbr-/- (n=5), LtbrΔFb (n=9), or LtbrΔTEC (n=5) mice. Significance was determined by one-way ANOVA with Holm-Sidak’s multiple comparison test. (d and e) Intercellular protein transfer from fibroblasts to thymic DCs. d, EGFP expression in thymic cDCs (CD11chi PDCA1lo) from control mice (loxP-EGFP) (n=5), fibroblast-specific EGFP-expressing mice (Twist2-Cre loxP-EGFP) (n=4), or TEC-specific EGFP-expressing mice (Foxn1-Cre loxP-EGFP) (n=5). e, T-cell-depleted bone marrow (BM) cells from B6-CD45.1 mice were transferred into lethally irradiated control mice (loxP-EGFP) (n=5), fibroblast-specific EGFP-expressing mice (Twist2-Cre loxP-EGFP) (n=5), or TEC-specific EGFP-expressing mice (Foxn1-Cre loxP-EGFP) (n=3). EGFP expression in donor-derived thymic cDCs (CD45.1+ CD11chi PDCA1lo) was analyzed 5 weeks after the transplantation. In d and e, significance was determined between control group and Twist2-Cre group by unpaired two-tailed t-test.
Extended Data Fig. 10 T cell development in LTβR-deficient mice.
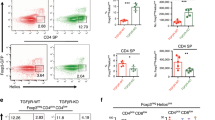
a, Representative flow cytometry profiles for CD4 and CD8 expression of whole thymocytes. b, Flow cytometry profiles for CD62L and CD69 expression of gated CD4+CD8-TCRβ+ (CD4SP) or CD4-CD8+TCRβ+ (CD8SP) cells. Graphs show frequencies of CD62LhiCD69lo cells in CD4SP or CD8SP cells. In a and b, data were obtained using Ltbrflox (n=10), Ltbr-/- (n=6), LtbrΔFb (n=7), or LtbrΔTEC (n=5) mice from ten independent experiments. Significance was determined by one-way ANOVA with Holm-Sidak’s multiple comparison test. c, Generation of TCRβ-retrogenic mice. Sca1+ bone marrow cells from TCRα+/- (Tcra+/-) mice were retrovirally transduced with TCRβ and transplanted into irradiated Ltbrflox (n=3) or LtbrΔFb mice (n=3). The mice were analyzed 5 weeks after the transplantation. Representative flow cytometry profiles of CD4 and CD8 expression in the GFP+ Vβ5hi thymocytes (left) and frequency of CD4SP or CD8SP cells in gated GFP+ Vβ5hi thymocytes (right). Significance was determined by unpaired two-tailed t-test.
Supplementary information
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical Source Data
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
About this article
Cite this article
Nitta, T., Tsutsumi, M., Nitta, S. et al. Fibroblasts as a source of self-antigens for central immune tolerance. Nat Immunol 21, 1172–1180 (2020). https://doi.org/10.1038/s41590-020-0756-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-020-0756-8
This article is cited by
-
The Proteostasis of Thymic Stromal Cells in Health and Diseases
The Protein Journal (2024)
-
Peripheral T-cell responses of EphB2- and EphB3-deficient mice in a model of collagen-induced arthritis
Cellular and Molecular Life Sciences (2024)
-
Nitric oxide-dependent immunosuppressive function of thymus-derived mesenchymal stromal/stem cells
Biology Direct (2023)
-
Potential role of mesenchymal stem cells in T cell aging
Journal of Molecular Medicine (2023)
-
Mesenchymal stromal cells in the thymus
Inflammation and Regeneration (2022)