Abstract
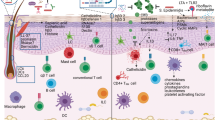
Skin wounds heal by coordinated induction of inflammation and tissue repair, but the initiating events are poorly defined. Here we uncover a fundamental role of commensal skin microbiota in this process and show that it is mediated by the recruitment and the activation of type I interferon (IFN)-producing plasmacytoid DC (pDC). Commensal bacteria colonizing skin wounds trigger activation of neutrophils to express the chemokine CXCL10, which recruits pDC and acts as an antimicrobial protein to kill exposed microbiota, leading to the formation of CXCL10–bacterial DNA complexes. These complexes and not complexes with host-derived DNA activate pDC to produce type I IFNs, which accelerate wound closure by triggering skin inflammation and early T cell–independent wound repair responses, mediated by macrophages and fibroblasts that produce major growth factors required for healing. These findings identify a key function of commensal microbiota in driving a central innate wound healing response of the skin.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the article and its supplementary information and from the corresponding author upon reasonable request. Source data are provided with this paper.
References
Sen, C. K. et al. Human skin wounds: a major and snowballing threat to public health and the economy. Wound Repair Regen. 17, 763–771 (2009).
Singer, A. J. & Clark, R. A. Cutaneous wound healing. New Engl. J. Med. 341, 738–746 (1999).
Martin, P. Wound healing—aiming for perfect skin regeneration. Science 276, 75–81 (1997).
Gurtner, G. C., Werner, S., Barrandon, Y. & Longaker, M. T. Wound repair and regeneration. Nature 453, 314–321 (2008).
Eming, S. A., Martin, P. & Tomic-Canic, M. Wound repair and regeneration: mechanisms, signaling, and translation. Sci. Transl. Med. 6, 265sr266 (2014).
Theofilopoulos, A. N., Baccala, R., Beutler, B. & Kono, D. H. Type I interferons (α/β) in immunity and autoimmunity. Ann. Rev. Immunol. 23, 307–336 (2005).
Sun, L. et al. Type I interferons link viral infection to enhanced epithelial turnover and repair. Cell Host Microbe. 17, 85–97 (2015).
Gregorio, J. et al. Plasmacytoid dendritic cells sense skin injury and promote wound healing through type I interferons. J. Exp. Med. 207, 2921–2930 (2010).
Fischer, J. C. et al. RIG-I/MAVS and STING signaling promote gut integrity during irradiation- and immune-mediated tissue injury. Sci. Transl. Med. 9, eaag2513 (2017).
Tohyama, M. et al. IFN-α enhances IL-22 receptor expression in keratinocytes: a possible role in the development of psoriasis. J. Invest. Dermatol. 132, 1933–1935 (2012).
Lande, R. et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature 449, 564–569 (2007).
Lande, R. et al. Cationic antimicrobial peptides in psoriatic skin cooperate to break innate tolerance to self-DNA. Euro. J. Immunol. 45, 203–213 (2015).
Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 415, 389–395 (2002).
Stacy, A. & Belkaid, Y. Microbial guardians of skin health. Science 363, 227–228 (2019).
Naik, S. et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature 520, 104–108 (2015).
Linehan, J. L. et al. Non-classical immunity controls microbiota impact on skin immunity and tissue repair. Cell 172, 784–796.e18 (2018).
Werner, S. & Grose, R. Regulation of wound healing by growth factors and cytokines. Physiol. Rev. 83, 835–870 (2003).
Bradley, K. C. et al. Microbiota-driven tonic interferon signals in lung stromal cells protect from influenza virus infection. Cell Rep. 28, 245–256.e4 (2019).
Martin, P. K. et al. Autophagy proteins suppress protective type I interferon signalling in response to the murine gut microbiota. Nat Microbiol. 3, 1131–1141 (2018).
Vanbervliet, B. et al. The inducible CXCR3 ligands control plasmacytoid dendritic cell responsiveness to the constitutive chemokine stromal cell-derived factor 1 (SDF-1)/CXCL12. J. Exp. Med. 198, 823–830 (2003).
Ordonez-Rueda, D. et al. A hypomorphic mutation in the Gfi1 transcriptional repressor results in a novel form of neutropenia. Euro. J. Immunol. 42, 2395–2408 (2012).
Cole, A. M. et al. Cutting edge: IFN-inducible ELR– CXC chemokines display defensin-like antimicrobial activity. J. Immunol. 167, 623–627 (2001).
Margulieux, K. R., Fox, J. W., Nakamoto, R. K. & Hughes, M. A. CXCL10 acts as a bifunctional antimicrobial molecule against Bacillus anthracis. MBio 7, e00334–16 (2016).
Bauer, S. et al. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc. Natl Acad. Sci. USA 98, 9237–9242 (2001).
Krieg, A. M. et al. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature 374, 546–549 (1995).
Stetson, D. B. & Medzhitov, R. Type I interferons in host defense. Immunity. 25, 373–381 (2006).
Jameson, J. M., Sharp, L. L., Witherden, D. A. & Havran, W. L. Regulation of skin cell homeostasis by gamma delta T cells. Front Biosci. 9, 2640–2651 (2004).
Kono, H. & Rock, K. L. How dying cells alert the immune system to danger. Nat. Rev. Immunol. 8, 279–289 (2008).
Yang, D. et al. Many chemokines including CCL20/MIP-3α display antimicrobial activity. J. Leukocyte Biol. 74, 448–455 (2003).
Lee, E. Y. et al. Helical antimicrobial peptides assemble into protofibril scaffolds that present ordered dsDNA to TLR9. Nat. Commun. 10, 1012 (2019).
Schmidt, N. W. et al. Liquid-crystalline ordering of antimicrobial peptide-DNA complexes controls TLR9 activation. Nat. Mater. 14, 696–700 (2015).
Wolf, M. & Moser, B. Antimicrobial activities of chemokines: not just a side-effect? Front. Immunol. 3, 213 (2012).
Nestle, F. O. et al. Plasmacytoid predendritic cells initiate psoriasis through interferon-α production. J. Exp. Med. 202, 135–143 (2005).
Stehlikova, Z. et al. Dysbiosis of skin microbiota in psoriatic patients: co-occurrence of fungal and bacterial communities. Front Microbiol. 10, 438 (2019).
De Cecco, M. et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566, 73–78 (2019).
Demaria, M. et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev. Cell. 31, 722–733 (2014).
Hiebert, P. et al. Nrf2-Mediated Fibroblast Reprogramming Drives Cellular Senescence by Targeting the Matrisome. Dev. Cell 46, 145–161.e10 (2018).
Jameson, J. et al. A role for skin γδ T cells in wound repair. Science 296, 747–749 (2002).
Harrison, O. J. et al. Commensal-specific T cell plasticity promotes rapid tissue adaptation to injury. Science 363, eaat6280 (2019).
Loesche, M. et al. Temporal stability in chronic wound microbiota is associated with poor healing. J. Invest. Dermatol. 137, 237–244 (2017).
Yankeelov, J. A. Jr. Modification of arginine in proteins by oligomers of 2,3-butanedione. Biochemistry. 9, 2433–2439 (1970).
Akbar, A. N. et al. Investigation of the cutaneous response to recall antigen in humans in vivo. Clin. Exp. Immunol. 173, 163–172 (2013).
Acknowledgements
We thank M. Malissen from INSERM U1104 laboratory in CIML, Marseille, France for providing the Genista mice; B. Humbel and J. Daraspe from the Electron Microscopy Facility of the University of Lausanne for the electron microscopy; H. Maby-El Hajjami, L. Cagnon, S.A. Maillard and F. Stuber for recruiting healthy volunteers and for establishing and running the human skin blister study; and I. Surbeck and A. Joncic for technical assistance. This work was funded by the Swiss National Science Foundation to M.G. (grant no. 310030_163360), a grant from the Fondation Pierre Mercier to J.D.D., and an Alfred and Annemarie von Sick grant to D.E.S. and M.G.
Author information
Authors and Affiliations
Contributions
J.D.D. and C.B. performed and analyzed most of the experiments and participated in their design. P.C., A.F. and B.R. helped establish and perform GF experiments. T.M., P.M.M. and D.E.S. designed and performed the skin blister induction experiments. O.D. and C.C. helped with some experiments and, along with S.W., contributed to discussions throughout the study. B.H. performed immunohistology for CXCL10. M.G. conceived and supervised the study, was involved in the design and evaluation of all experiments and wrote the manuscript along with J.D.D. and C.B., with comments from coauthors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Zoltan Fehervari was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 CXCL10 is expressed by skin-infiltrating neutrophils.
Images of histological sections of injured skins harvested before (0h) or 6, 12, and 24 hours postinjury. Sections were stained by chromogenic immunohistochemistry using DAB with control isotypes, anti-CXCL10, or anti-Ly6G antibodies. Skin sections were then counterstained with hematoxylin. Images are representative of 5 different mice. Scale bars are shown and represent 50 μM.
Extended Data Fig. 2 Accumulation of non-hemolytic catalase-positive cocci in the dermis of tape-stripped skin.
a FISH staining of bacterial 16S rRNA of uninjured (top) and tape-stripped (bottom) murine skin. Reverse probes for 16S rRNA were used as controls. Scale bars are shown on images. b, Colorimetric Gram staining of uninjured and injured skin, showing the presence of cocci in the dermis of stripped skin but not in uninjured skin. a-c, images are representative of 5 different mice. c, Photographs of a blood-agar plate showing non-hemolytic bacterial colonies grown overnight from tape-stripped skin of three mice (top line). One colony per mouse was tested for catalase activity by adding 3% H2O2 . Absence of gas bubbles indicates catalase negativity (middle line). Gram staining of bacteria harvested from the injured skin analyzed by light microscopy (bottom line).
Extended Data Fig. 3 Topical Neosporin ointment treatment efficiently reduces commensal bacteria in murine skin.
a, Photographs of a blood-agar plate showing bacterial colonies grown overnight from skin samples harvested from five different vehicle-treated and Neosporin-treated mice (left). The number of colony forming units (CFU) in each group was then calculated (right). Data represent the mean + s.e.m. n = 5. *P<0.01, **P<0.001 (one-way ANOVA followed by Tukey’s multiple comparisons test). b, Quantification of bacterial 16S rDNA by qPCR in the skin of vehicle-treated, Neosporin-treated, or Neosporin-treated Staphylococcus epidermidis-recolonized SPF-housed mice, and untreated or Staphylococcus epidermidis-recolonized germ-free (GF) mice.
Extended Data Fig. 4 Only CXCL10 skin injection induces Ifna, but all CXCL9, CXCL10, and CXCL11 kill bacteria, bind DNA, and induce type I IFN production in pDC.
a, Expression of Ifna2 in the skin of mice 6 hours after intradermal injection of CXCL9, CXCL10, CXCL11, chemerin, or control saline. Data represent the mean + s.e.m. n = 4. *P<0.01 (one-way ANOVA followed by Tukey’s multiple comparisons test). b, Percentages of viable SytoGreen+ SytoxOrange- bacteria (Staphylococcus epidermidis) incubated for 24 hours with increasing concentrations of CXCL9, CXCL10, CXCL11, or Chemerin assessed by flow cytometry. Data represent the mean + sd of triplicates. c, Fluorimetric quantification of DNA staining by PicoGreen® dye after mixing of bacterial DNA with CXCL9, CXCL10, CXCL11, or Chemerin. A.U., arbitrary units. Data represent the mean + sd of triplicates. * P<0.05 (one-way ANOVA followed by Dunnett’s multiple comparisons test). d, Amounts of IFN-α produced by human blood-isolated pDC stimulated for 24 hours with purified DNA from bacteria in the presence or not of CXCL9, CXCL10, CXCL11, or Chemerin. Data represent the mean + sd. n=3. **** P<0.0001 (two-way ANOVA followed by Sidak’s multiple comparisons test).
Extended Data Fig. 5 Intradermal injection leads to bacterial translocation from the epidermis into the dermis.
a, Colorimetric Gram staining of murine skin 1 hour after intradermal injection of 50 ml of CXCL10-containing solution with a 30G needle showing bacterial cocci translocated into the dermis. b, FISH staining of bacterial 16S rRNA gene of the same intradermally injected murine skin than in (a). Reverse probes for 16S rRNA gene were used as controls. a-b, images are representative of 5 different mice. Scale bars are shown on images.
Extended Data Fig. 6 Expression of inflammatory and repair response genes in tape-stripped vs full-thickness skin injury.
Expression of Il1b, Ifna2, Cxcl10, Il6, Il12a, Il22, Il17a, Il23a, Il10, Tnf, Fgf2, Fgf7, Tgfb1, Vegfa, Pdgf, Ctgf, Egf, Il24, and Ifng genes in the uninjured or injured skin (tape-stripping or full thickness injury) at 12, 24, and 48 hours after skin injury assessed by RT-qPCR. Data are from five different mice per group and presented as a heatmap of fold changes. Data are representative of four independent experiments.
Extended Data Fig. 7 Neosporin treatment delays the wound closure rate, which is restored by re-colonizing mice with S.epidermidis or intradermal injection of IFN-α.
a, Photographs of skin wounds in untreated (vehicle), Neosporin-treated, Staphylococcus epidermidis-recolonized Neosporin-treated, and IFNα-intradermally injected Neosporin treated mice at the time of wounding (0) and everyday up to 6 days post-injury. b, Wound closure kinetics shown as percentages of initial wound size in the different mice groups as in (a). Data are the mean±SD of three mice per group are representative of 2 independent experiments. Scale bars are shown on images.
Extended Data Fig. 8 Skin injury activates dermal cells to produce growth factors.
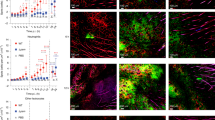
a, Confocal microscopy images of FFPE murine skin sections at 0 (before) and 48 hours after injury (bottom) stained with DAPI (blue), and antibodies against vimentin (green), and FGF2 (red) or with isotype control (red). Images shown are representative of three different mice from two independent experiments. Scale bars shown on images represent 50 μm. b, Confocal microscopy images of frozen murine skin sections at 0 (before) and 48 hours after injury (bottom) stained with DAPI (blue), and antibodies against CD206 (green), and TGFb (red) or with isotype control (red). Images shown are representative of three different mice from two independent experiments. Scale bars shown on images represent 50 μm.
Supplementary information
Source data
Source Data Fig. 1
Source data
Source Data Fig. 2
Source data
Source Data Fig. 3
Source data
Source Data Fig. 7
Source data
Rights and permissions
About this article
Cite this article
Di Domizio, J., Belkhodja, C., Chenuet, P. et al. The commensal skin microbiota triggers type I IFN–dependent innate repair responses in injured skin. Nat Immunol 21, 1034–1045 (2020). https://doi.org/10.1038/s41590-020-0721-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-020-0721-6
This article is cited by
-
Cellular and molecular mechanisms of skin wound healing
Nature Reviews Molecular Cell Biology (2024)
-
The efficiency of zinc sulfate immersion bath on improved wound healing via promoting antioxidant activity, gene expression biomarkers, and skin re-epithelization in a common carp-induced wound model
Applied Water Science (2024)
-
Skin Deep: The Potential of Microbiome Cosmetics
Journal of Microbiology (2024)
-
Staphylococcus epidermidis and its dual lifestyle in skin health and infection
Nature Reviews Microbiology (2023)
-
Controlling skin microbiome as a new bacteriotherapy for inflammatory skin diseases
Inflammation and Regeneration (2022)