Abstract
The molecular basis for the propensity of a small number of environmental proteins to provoke allergic responses is largely unknown. Herein, we report that mite group 13 allergens of the fatty acid-binding protein (FABP) family are sensed by an evolutionarily conserved acute-phase protein, serum amyloid A1 (SAA1), that promotes pulmonary type 2 immunity. Mechanistically, SAA1 interacted directly with allergenic mite FABPs (Der p 13 and Blo t 13). The interaction between mite FABPs and SAA1 activated the SAA1-binding receptor, formyl peptide receptor 2 (FPR2), which drove the epithelial release of the type-2-promoting cytokine interleukin (IL)-33 in a SAA1-dependent manner. Importantly, the SAA1–FPR2–IL-33 axis was upregulated in nasal epithelial cells from patients with chronic rhinosinusitis. These findings identify an unrecognized role for SAA1 as a soluble pattern recognition receptor for conserved FABPs found in common mite allergens that initiate type 2 immunity at mucosal surfaces.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Wills-Karp, M., Nathan, A., Page, K. & Karp, C. L. New insights into innate immune mechanisms underlying allergenicity. Mucosal Immunol. 3, 104–110 (2010).
Corrigan, C. J. et al. Allergen-induced expression of IL-25 and IL-25 receptor in atopic asthmatic airways and late-phase cutaneous responses. J. Allergy Clin. Immunol. 128, 116–124 (2011).
Gour, N. et al. Dysregulated invertebrate tropomyosin–dectin-1 interaction confers susceptibility to allergic diseases. Sci. Immunol. 3, eaam9841 (2018).
Schuijs, M. J. et al. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science 349, 1106–1110 (2015).
Smole, U., Kratzer, B. & Pickl, W. F. Soluble pattern recognition molecules: guardians and regulators of homeostasis at airway mucosal surfaces. Eur. J. Immunol. 50, 624–642 (2020).
Urieli-Shoval, S., Cohen, P., Eisenberg, S. & Matzner, Y. Widespread expression of serum amyloid A in histologically normal human tissues. Predominant localization to the epithelium. J. Histochem. Cytochem. 46, 1377–1384 (1998).
Malle, E., Sodin-Semrl, S. & Kovacevic, A. Serum amyloid A: an acute-phase protein involved in tumour pathogenesis. Cell. Mol. Life Sci. 66, 9–26 (2009).
Cai, Z. et al. Human serum amyloid A protein inhibits hepatitis C virus entry into cells. J. Virol. 81, 6128–6133 (2007).
Shah, C., Hari-Dass, R. & Raynes, J. G. Serum amyloid A is an innate immune opsonin for Gram-negative bacteria. Blood 108, 1751–1757 (2006).
de Beer, M. C. et al. Impact of serum amyloid A on high density lipoprotein composition and levels. J. Lipid Res. 51, 3117–3125 (2010).
Hinrichs, B. H. et al. Serum amyloid A1 is an epithelial prorestitutive factor. Am. J. Pathol. 188, 937–949 (2018).
Sun, L. & Ye, R. D. Serum amyloid A1: structure, function and gene polymorphism. Gene 583, 48–57 (2016).
Cheng, N., Liang, Y., Du, X. & Ye, R. D. Serum amyloid A promotes LPS clearance and suppresses LPS-induced inflammation and tissue injury. EMBO Rep. 19, e45517 (2018).
Kim, M. H., de Beer, M. C., Wroblewski, J. M., Webb, N. R. & de Beer, F. C. SAA does not induce cytokine production in physiological conditions. Cytokine 61, 506–512 (2013).
Lu, J., Yu, Y., Zhu, I., Cheng, Y. & Sun, P. D. Structural mechanism of serum amyloid A-mediated inflammatory amyloidosis. Proc. Natl Acad. Sci. USA 111, 5189–5194 (2014).
Wang, Y. et al. Serum amyloid A 2.2 refolds into a octameric oligomer that slowly converts to a more stable hexamer. Biochem. Biophys. Res. Commun. 407, 725–729 (2011).
Sano, T. et al. An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell 163, 381–393 (2015).
Ather, J. L. et al. Serum amyloid A activates the NLRP3 inflammasome and promotes Th17 allergic asthma in mice. J. Immunol. 187, 64–73 (2011).
Buyukozturk, S. et al. Acute phase reactants in allergic airway disease. Tohoku J. Exp. Med. 204, 209–213 (2004).
Jousilahti, P. et al. The association of sensitive systemic inflammation markers with bronchial asthma. Ann. Allergy Asthma Immunol. 89, 381–385 (2002).
Ozseker, F. et al. Serum amyloid A (SAA) in induced sputum of asthmatics: a new look to an old marker. Int. Immunopharmacol. 6, 1569–1576 (2006).
Ricklefs, I. et al. ALX receptor ligands define a biochemical endotype for severe asthma. JCI Insight 2, e93534 (2017).
Lambrecht, B. N., Hammad, H. & Fahy, J. V. The cytokines of asthma. Immunity 50, 975–991 (2019).
Cayrol, C. et al. Environmental allergens induce allergic inflammation through proteolytic maturation of IL-33. Nat. Immunol. 19, 375–385 (2018).
Gold, M. J. et al. Group 2 innate lymphoid cells facilitate sensitization to local, but not systemic, TH2-inducing allergen exposures. J. Allergy Clin. Immunol. 133, 1142–1148 (2014).
Halim, T. Y. et al. Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity 40, 425–435 (2014).
Ather, J. L. et al. Serum amyloid A3 is required for normal lung development and survival following influenza infection. Sci. Rep. 8, 16571 (2018).
Pautsch, A. & Schulz, G. E. High-resolution structure of the OmpA membrane domain. J. Mol. Biol. 298, 273–282 (2000).
Niemi, M. H. et al. Dimerization of lipocalin allergens. Sci. Rep. 5, 13841 (2015).
Malle, E. et al. Mapping of antigenic determinants of purified, lipid-free human serum amyloid A proteins. Scand. J. Immunol. 48, 557–561 (1998).
Suojalehto, H. et al. Level of fatty acid binding protein 5 (FABP5) is increased in sputum of allergic asthmatics and links to airway remodeling and inflammation. PLoS ONE 10, e0127003 (2015).
Thaumaturgo, N., Vilar, M. M., Diogo, C. M., Edelenyi, R. & Tendler, M. Preliminary analysis of Sm14 in distinct fractions of Schistosoma mansoni adult worm extract. Mem. Inst. Oswaldo Cruz 96 (Suppl. I), 79–83 (2001).
Franchini, G. R. et al. The unusual lipid binding proteins of parasitic helminths and their potential roles in parasitism and as therapeutic targets. Prostaglandins Leukot. Essent. Fatty Acids 93, 31–36 (2015).
Derebe, M. G. et al. Serum amyloid A is a retinol binding protein that transports retinol during bacterial infection. Elife 3, e03206 (2014).
Zhou, H., Chen, M., Zhang, G. & Ye, R. D. Suppression of lipopolysaccharide-induced inflammatory response by fragments from serum amyloid A. J. Immunol. 199, 1105–1112 (2017).
Rabiet, M. J., Macari, L., Dahlgren, C. & Boulay, F. N-formyl peptide receptor 3 (FPR3) departs from the homologous FPR2/ALX receptor with regard to the major processes governing chemoattractant receptor regulation, expression at the cell surface, and phosphorylation. J. Biol. Chem. 286, 26718–26731 (2011).
Cooray, S. N. et al. Ligand-specific conformational change of the G-protein-coupled receptor ALX/FPR2 determines proresolving functional responses. Proc. Natl Acad. Sci. USA 110, 18232–18237 (2013).
Snelgrove, R. J. et al. Alternaria-derived serine protease activity drives IL-33-mediated asthma exacerbations. J. Allergy Clin. Immunol. 134, 583–592 (2014).
Green, B. J. et al. Allergic sensitization in Canadian chronic rhinosinusitis patients. Allergy Asthma Clin. Immunol. 10, 15 (2014).
Chu, D. K. et al. IL-33, but not thymic stromal lymphopoietin or IL-25, is central to mite and peanut allergic sensitization. J. Allergy Clin. Immunol. 131, 187–200 (2013).
Rank, M. A. et al. IL-33–activated dendritic cells induce an atypical TH2-type response. J. Allergy Clin. Immunol. 123, 1047–1054 (2009).
Besnard, A. G. et al. IL-33-activated dendritic cells are critical for allergic airway inflammation. Eur. J. Immunol. 41, 1675–1686 (2011).
Frame, N. M., Jayaraman, S., Gantz, D. L. & Gursky, O. Serum amyloid A self-assembles with phospholipids to form stable protein-rich nanoparticles with a distinct structure: a hypothetical function of SAA as a “molecular mop” in immune response. J. Struct. Biol. 200, 293–302 (2017).
Jayaraman, S., Gantz, D. L., Haupt, C. & Gursky, O. Serum amyloid A forms stable oligomers that disrupt vesicles at lysosomal pH and contribute to the pathogenesis of reactive amyloidosis. Proc. Natl Acad. Sci. USA 114, E6507–E6515 (2017).
Bjorkman, L. et al. The proinflammatory activity of recombinant serum amyloid A is not shared by the endogenous protein in the circulation. Arthritis Rheum. 62, 1660–1665 (2010).
Christenson, K. et al. Endogenous acute phase serum amyloid A lacks pro-inflammatory activity, contrasting the two recombinant variants that activate human neutrophils through different receptors. Front. Immunol. 4, 92 (2013).
Puerta, L., Kennedy, M. W., Jimnez, S. & Caraballo, L. Structural and ligand binding analysis of recombinant Blo t 13 allergen from Blomia tropicalis mite, a fatty acid binding protein. Int. Arch. Allergy Immunol. 119, 181–184 (1999).
Scott, I. C. et al. Interleukin-33 is activated by allergen- and necrosis-associated proteolytic activities to regulate its alarmin activity during epithelial damage. Sci. Rep. 8, 3363 (2018).
Ivanov, I. I. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
Caraballo, L. et al. Cloning and IgE binding of a recombinant allergen from the mite Blomia tropicalis, homologous with fatty acid-binding proteins. Int. Arch. Allergy Immunol. 112, 341–347 (1997).
Zimmerman, A. W. & Veerkamp, J. H. New insights into the structure and function of fatty acid-binding proteins. Cell. Mol. Life Sci. 59, 1096–1116 (2002).
Miljkovic, D. et al. Association between group 2 innate lymphoid cells enrichment, nasal polyps and allergy in chronic rhinosinusitis. Allergy 69, 1154–1161 (2014).
Shaw, J. L. et al. IL-33-responsive innate lymphoid cells are an important source of IL-13 in chronic rhinosinusitis with nasal polyps. Am. J. Respir. Crit. Care Med. 188, 432–439 (2013).
Chen, M., Zhou, H., Cheng, N., Qian, F. & Ye, R. D. Serum amyloid A1 isoforms display different efficacy at Toll-like receptor 2 and formyl peptide receptor 2. Immunobiology 219, 916–923 (2014).
Lotvall, J. et al. Asthma endotypes: a new approach to classification of disease entities within the asthma syndrome. J. Allergy Clin. Immunol. 127, 355–360 (2011).
Leontovyc, A. et al. SmSP2: a serine protease secreted by the blood fluke pathogen Schistosoma mansoni with anti-hemostatic properties. PLoS Negl. Trop. Dis. 12, e0006446 (2018).
Lajoie, S. et al. IL-21 receptor signalling partially mediates Th2-mediated allergic airway responses. Clin. Exp. Allergy 44, 976–985 (2014).
Paris, G., Pozharskaya, T., Asempa, T. & Lane, A. P. Damage-associated molecular patterns stimulate interleukin-33 expression in nasal polyp epithelial cells. Int. Forum Allergy Rhinol. 4, 15–21 (2014).
Darveau, M. E., Jacques, E., Rouabhia, M., Hamid, Q. & Chakir, J. Increased T-cell survival by structural bronchial cells derived from asthmatic subjects cultured in an engineered human mucosa. J. Allergy Clin. Immunol. 121, 692–699 (2008).
Goulet, F. et al. Morphologic and functional properties of bronchial cells isolated from normal and asthmatic subjects. Am. J. Respir. Cell Mol. Biol. 15, 312–318 (1996).
Labrada, M. et al. Monoclonal antibodies against Blo t 13, a recombinant allergen from Blomia tropicalis. Int. Arch. Allergy Immunol. 129, 212–218 (2002).
Kratzer, B. et al. Prevention of allergy by virus-like nanoparticles (VNP) delivering shielded versions of major allergens in a humanized murine allergy model. Allergy 74, 246–260 (2019).
Acknowledgements
We thank M. C. de Beer (University of Kentucky Medical Center, KY, US) for providing the Saa−/− mice; E. Schmidt and R. Zeiner for animal care; and Z. Lijie, D. Trapin and S. Kickmaier for expert technical assistance. We also thank M. Marlovics (dsgn&cde; hq@dsgncde.com) for the graphic designs in Figs. 5 and 8 and C. Zwicker for critically reading the manuscript and for helpful discussion and suggestions. This work was funded by the National Institute of Allergy and Infectious Diseases (grants U19AI070235 and R01 AI083315 to M.W.-K.) and the NIH (grants R56AI118791 and R01AI127644 to S.L. and R01AI132590 to A.P.L.) as well as the Austrian Science Fund (DK W1248 and SFB F4609 to W.F.P.). U.S. was supported by an Erwin Schrödinger Fellowship (J3332-B21) of the Austrian Science Fund, a research grant of the American Thoracic Society and DK W1248. N.G. was supported by an NIEHS 5T32ES007141 grant. E.M. was supported by the Austrian National Bank (17600). J.D. was supported by Operational Programme Research, Development and Education, the Call International Mobility of Researchers – MSCA – IF (CZ.02.2.69/0.0/0.0/17_050/0008014). J.C. was supported by the Canadian Institutes of Health Research (DC0190GP).
Author information
Authors and Affiliations
Contributions
U.S., S.L. and M.W.-K. designed the study. U.S. and S.L. performed the experiments and analyzed the data with help from N.G., X.X., J.P., N.Y. and W.F.P. C.K., B.K. and P.A.T. performed experiments and provided technical assistance with the allergic phenotype involving Saa−/− mice. G.H. produced and characterized rSAA1.1. J.D. provided S. mansoni worm extracts. L.C. and L.P. made B. tropicalis rFABP and B. tropicalis FABP-specific monoclonal antibodies. E.M. provided peptide-specific SAA antibodies and performed SAA1 antibody blots. S.R. provided serum samples from control individuals and patients with a HDM allergy. A.P.L. provided samples from control individuals and patients with CRS. J.C. isolated and prepared AECs from controls and individuals with asthma. W.F.P. and E.M. thoroughly revised the manuscript. U.S. and M.W.-K. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information L. A. Dempsey was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Experimental design.
a, Model of allergen-induced airway hyperresponsiveness (AHR) for wild-type (WT) and Saa–/– mice (both C57BL/6 background). Mice were sensitized i.t. on day 0 (1 μg) and i.n. on days 7-11 with 10 μg of HDM extract. Airway measurements were performed 72 h after the last allergen challenge (used in Fig. 1a–j and Extended Data Figs. 2 and 3). b, For SAA1 antibody blockade, we used an established mouse model of allergen-induced AHR sensitizing WT BALB/cJ mice i.t. on day 0 and 14 with 100 µg of HDM extract + isotype control, or HDM + anti-SAA. Airway measurements and tissue harvests were performed 72 h after the last allergen challenge (used in Extended Data Fig. 4). c, In short-term exposure protocols WT and Saa–/– mice (both C57BL/6 background) received a single HDM challenge (100 μg) were sacrificed 16 h later. d, In short-term exposure protocols BALB/cJ mice received a single HDM challenge (100 μg) + isotype control, HDM challenge + HDL (200μg) and isotype control, or HDM + anti-SAA and were sacrificed 24 h later (used in Fig. 2e). e, For overexpression of SAA1 in vivo, mice were injected 20 µg of DNA complexed to polyethylenimine at day 0, exposed to PBS or HDM 48 h later and ILC2s as well as BAL cytokines measurements were performed on day 3 (used Fig. 2f). f, WT and Saa−/− mice were sensitized i.t. on day 0 (1 μg) and i.n. on days 7-11 with 10 μg with extracts from the parasitic worm Schistosoma mansoni (a Puerto Rican isolate). Tissues were harvested 72 h after the last allergen challenge (used in Fig. 4). g, For FPR2 blockade (WRW4, 2 mg/kg), WT BALB/cJ mice were sensitized and challenged i.t. on day 0 and 14 with 100 μg of HDM extract. Airway measurements were performed 72 h after the last allergen challenge (used in Fig. 7a–f). h, In short term exposure experiments, BALB/cJ mice received a single HDM challenge (100 μg) or HDM + WRW4 and were sacrificed 24 h later (used in Fig. 7g, h). i, Model of A. alternata (Alt a)-induced airway inflammation for WT and Saa–/– mice. Mice were sensitized i.t. on day 0 (1 μg) and i.n. on days 7-11 with 10 µg of Alt a extract. Tissues were harvested 72 h after the last allergen challenge (used in Extended Data Fig. 9f–j).
Extended Data Fig. 2 Reduced recruitment of CD11b+ DCs in Saa–/– mice after allergen exposure.
Allergic phenotype in WT and Saa–/– C57BL/6 mice sensitized and challenged with PBS or HDM was analyzed seventy-two hours after the last challenge. a, Lung cell gating strategy used in Fig. 1, Fig. 4 and Extended Data Figs. 6 and 9. Frequency of dendritic cell (DC) populations (b-d), monocytes (e-f) and granulocytes (g-h) in the lungs of WT and Saa–/– mice used in Fig. 1. Data represent means ± s.e.m. of pooled data from 2 independent experiments containing n=11 WT PBS, n=12 WT HDM, n=11 Saa–/– PBS and n=13 Saa–/– HDM animals per group. P values were calculated with a two-tailed test using one-way analysis of variance (ANOVA) with Dunett’s post hoc analysis that compares WT HDM to counterparts. **P = 0.0039, ****P ≤ 0.0001.
Extended Data Fig. 3 Reduced migration of CD3+CD4+ T cells to the lungs of Saa–/– mice after allergen exposure.
Allergic phenotype in WT and Saa–/– C57BL/6 mice sensitized and challenged with PBS or HDM was analyzed seventy-two hours after the last challenge. a, ILC and T cell gating strategy (used in Fig. 1g, h, Fig. 4b–e and Extended Data Fig. 9h–j). b, Total lung cell counts (**P = 0.0073), (c) frequency and (d) numbers of CD3+CD4+ T cells (**P = 0.0014) and (e) frequency and numbers of (f) ICOS+ST2+ ILCs in the lungs of WT and Saa–/– mice. Data represent pooled data presented as means ± s.e.m. from 2 independent experiments containing n=11 WT PBS, n= 12 WT HDM, n=11 Saa–/– PBS and n=13 Saa–/– HDM animals per group. P values were calculated with a two-tailed test using one-way analysis of variance (ANOVA) with Dunett’s post hoc analysis that compares WT HDM to counterparts.
Extended Data Fig. 4 Airway SAA1-neutralization ameliorates the allergic phenotype.
a, AHR (*P = 0.0105), (b) total serum IgE concentrations (*P = 0.0265), (c) eosinophil infiltration into the lungs, and (d) PAS stained lung sections of isotype (iso), HDM+isotype (HDM), or HDM+SAAab-treated (anti-SAA) WT BALB/c mice. Antibodies were administered at 5 μg/i.t. Frequency of (e) Lin−CD45+ST2+IL-13+ ILC2s (**P = 0.0027, ***P = 0.0002), (f) TH2 and (*P = 0.0260, ***P = 0.0001) (g) TH17 cells (*P = 0.0149, ***P = 0.0005) in the lungs of these mice. Data represents means ± s.e.m. of pooled data from 2 independent experiments containing (a) n=6 PBS+iso, n=7 HDM+iso, n=9 HDM+anti-SAA animals per group; (b, c, f, g) n=9 PBS+iso, n=11 HDM+iso, n=13 HDM+anti-SAA animals per group or are representative of 2 independent experiments with (e) n=4 PBS+iso, n=5 HDM+iso, n=7 HDM+anti-SAA animals per group. P values were calculated with a two-tailed test using one-way analysis of variance (ANOVA) with Dunett’s post hoc analysis that compares HDM to iso and anti-SAA counterparts. ****P ≤ 0.0001.
Extended Data Fig. 5 SAA1 is a pattern recognition molecule for mite-derived and human cytosolic FABPs.
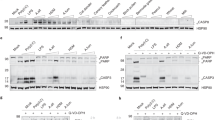
a, Migration of SAA1 (1 mg/ml) in IMDM media was analyzed in the presence of the mite FABP rBlo t 13 (1 mg/ml) by native PAGE followed by immunoblot analysis using a sequence-specific antiserum (amino acid 89-104) raised against human SAA1. b, Effects of the protein synthesis inhibitor cycloheximide on IL-33 concentrations in BEAS-2B cells treated for 30 min with HDM (100 μg/ml). c, Cell viability of BEAS-2B cells after HDM exposure over time as measured by continuous reduction of a cell viability substrate by viable cells (**P = 0.0041). SAA1 concentrations in cells after (d) siRNA-mediated silencing of SAA1 (siSAA1) or non-targeting scrambled siRNA (siNT). Effects of SAA1 on HDM-induced IL-6 (**P = 0.0086, ***P = 0.0007) and IL-8 release in cells with siRNA mediated knockdown of SAA1 (siSAA1) (e and f). g, HDM-induced IL-6 amounts after Der p 13-depletion and/or neutralization using a monoclonal antibody specific for mite group 13 allergens (anti-group 13). Data are shown as means ± s.e.m. and are representative of 2 (b, e) or 3 (c) independent experiments or pooled data from 2 independent experiments (d, f) each containing at least n=4 biologically independent samples. Representative analysis of SAA1 migration patterns in the presence of Blo t 13 using sequence-specific rabbit antiserum raised against human SAA1 (aa 89-104) (a). IL-6 amounts were analysed for one representative experiments with n=5 biologically independent samples (g). Cropped images are shown. P values were calculated with a two-tailed test using one-way analysis of variance (ANOVA) followed by Dunett’s post (b) or Tukey’s hoc analysis (e,f), two-way ANOVA followed by Dunnet correction (c) or Student’s t-test (d, g). ***P ≤ 0.001; ****P ≤ 0.0001.
Extended Data Fig. 6 Der p 13-depleted HDM has decreased TH2 skewing capacity.
a, Total IgE serum concentrations and numbers of BAL (b) eosinophils and (c) T cells in WT BALB/c mice undergoing a full allergen-exposure protocol using HDM that was depleted using either an isotype control antibody or a monoclonal antibody specific for mite group-13 allergens. d, Numbers of mediestinal lymph node IL-13+ CD4+ T cells (n.d., not detected) and (e) lung ILC2 (*P = 0.029). f-g, Cytokine production from HDM-restimulated lung cells (*P = 0.024). Data represent mean ± s.e.m. of n=2 PBS, n=4 HDM-isotype depleted, n=4 HDM- anti-group 13-depleted comparing HDM-isotype to α-group 13-depleted extract group using two-tailed unpaired t-test.
Extended Data Fig. 7 HDM-induced IL-33 is dependent on SAA1 dissociation.
a, mRNA and (b) protein amounts of SAA in response to HDM (100 μg/ml). c, SAA1 hexamer after rBlo t 13 stimulation was analyzed as described in Fig. 5b. d, Bar graph represents quantitative analysis of SAA hexamer using LI-COR Image Studio Software. e, Concentration-dependent IL-33 release from BEAS-2B cells induced by rBlo t 13. Data are shown as means ± s.e.m. and are pooled data from 2 independent experiments (b, e) each containing n=4-5 replicate wells or representative of 2-3 independent experiments (d). SAA1 mRNA expression, normalized to the average of housekeeping genes, is presented as mean value ± s.e.m. (n = 5). Immunoblot is representative of an experimental n=2. Cropped images are shown. P values were calculated with a two-tailed test using Student’s t-test (a), two-way analysis of variance (ANOVA) followed by Dunnet’s correction (b) or one-way ANOVA with Dunett’s post hoc analysis (d, e). ****P ≤ 0.0001.
Extended Data Fig. 8 The SAA1-FPR2 axis regulates IL-33.
a, mRNA expression of the FPR family members FPR1, FPR2 and FPR3 at baseline (filled bars) or after 2 h of HDM stimulation (white bars). b, HDM-triggered IL-33 amounts in BEAS-2B cells overexpressing human FPR1 (**P = 0.0068, ***P = 0.0003). c, IL-33 secretion in BEAS-2B cells overexpressing human FPR2 or cells transfected with an empty vector (EV; pcDNA3.1) (**P = 0.0028). HDM-induced IL-6 and IL-8 amounts in BEAS-2B cells overexpressing FPR2 (d and e) or blocking the FPR2 receptor f (**P = 0.0043) and g (**P = 0.0021)) using WRW4. Data presented as means ± s.e.m. and is representative of 2 independent experiments each containing at least n= 4 biologically independent samples (b, d, f) or pooled data from 2 independent experiments (c, e, g). mRNA expression, normalized to the average of housekeeping genes, is presented as mean values ± s.e.m. (n = 5 biologically independent samples) performed in duplicates (a). P values were calculated with a two-tailed test using Student’s t-test (a) or one-way analysis of variance (ANOVA) with Tukeys multiple comparison test (b-e) or Dunett’s post hoc analysis (f, g). ****P ≤ 0.0001.
Extended Data Fig. 9 SAA1-IL-33 axis is specific to HDM and not induced by A. alternata.
a, Immunoblot of SAA1 after A. alternata (Alt a) stimulation of BEAS-2B cells performed as described in Fig. 5b. Alt a-induced IL-33 and IL-8 (**P = 0.0044) secretion in BEAS-2B cells after siRNA-mediated silencing of SAA1 (siSAA1) (b and c) or WRW4-mediated FPR2 blockade (d and e). f, Total serum IgE concentrations, (g) eosinophil numbers and frequency of (h) CD3+CD4+, (i) TH2 and (j) TH17 cells in the lungs of PBS or Alt a-treated WT and Saa–/– C57BL/6 mice. Immunoblots are representative of an experimental n=2 (a). Data are presented as means ± s.e.m. and represent pooled data from 2 independent experiments (b, d, e, f, g, i, j) or show one representative experiment (c) each containing at least n= 4 biologically independent samples or n=8 WT PBS, n=13 WT Alt a, n=8 Saa–/– PBS and n=11 Saa–/– Alt a animals per group (f, g, i, j) or n=5 WT PBS, n=9 WT Alt a, n=5 Saa–/– PBS and n=7 Saa–/– Alt a animals per group (h). Cropped images are shown. P values were calculated with a two-tailed test using one-way analysis of variance (ANOVA) with Tukeys multiple comparison test (a, b) or Dunett’s post hoc analysis (d-j). ****P ≤ 0.0001 siNT = non-targeting siRNA; siSAA1 = SAA1-targeting siRNA. ns=not significant.
Extended Data Fig. 10 Dysregulated SAA and FPR2 expression in patients with asthma.
Basal (a) SAA1 and (b) FPR2 expression in bronchial epithelial cells from individuals with asthma and matched controls. Data represents means ± s.e.m. of (a and b) n= 6 control and n= 6 individuals with asthma per group. P values were calculated with a two-tailed test using Student’s t-test with Welch’s correction.
Supplementary information
Supplementary Information
Supplementary Tables 1–6.
Source data
Source Data Fig. 3
Unprocessed immunoblots of Fig. 3.
Source Data Fig. 5
Unprocessed immunoblots of Fig. 5.
Source Data Fig. 8
Unprocessed immunoblots of Fig. 8.
Source Data Extended Data Fig. 5
Unprocessed immunoblots of Extended Data Fig. 5.
Source Data Extended Data Fig. 7
Unprocessed immunoblots of Extended Data Fig. 7.
Source Data Extended Data Fig. 9
Unprocessed immunoblots of Extended Data Fig. 9.
Rights and permissions
About this article
Cite this article
Smole, U., Gour, N., Phelan, J. et al. Serum amyloid A is a soluble pattern recognition receptor that drives type 2 immunity. Nat Immunol 21, 756–765 (2020). https://doi.org/10.1038/s41590-020-0698-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-020-0698-1
This article is cited by
-
SAA suppresses α-PD-1 induced anti-tumor immunity by driving TH2 polarization in lung adenocarcinoma
Cell Death & Disease (2023)
-
Amnion-derived serum amyloid A1 participates in sterile inflammation of fetal membranes at parturition
Inflammation Research (2023)
-
Molecular recognition of formylpeptides and diverse agonists by the formylpeptide receptors FPR1 and FPR2
Nature Communications (2022)
-
High-Density Lipoproteins and Serum Amyloid A (SAA)
Current Atherosclerosis Reports (2021)
-
Purification and characterisation of the dimeric group 12 allergen from Blomia tropicalis heterologously expressed by Escherichia coli Top10F´
Molecular Biology Reports (2021)