Abstract
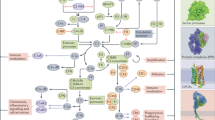
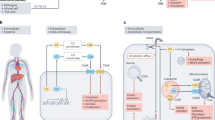
Complement hyperactivation, angiopathic thrombosis and protein-losing enteropathy (CHAPLE disease) is a lethal disease caused by genetic loss of the complement regulatory protein CD55, leading to overactivation of complement and innate immunity together with immunodeficiency due to immunoglobulin wasting in the intestine. We report in vivo human data accumulated using the complement C5 inhibitor eculizumab for the medical treatment of patients with CHAPLE disease. We observed cessation of gastrointestinal pathology together with restoration of normal immunity and metabolism. We found that patients rapidly renormalized immunoglobulin concentrations and other serum proteins as revealed by aptamer profiling, re-established a healthy gut microbiome, discontinued immunoglobulin replacement and other treatments and exhibited catch-up growth. Thus, we show that blockade of C5 by eculizumab effectively re-establishes regulation of the innate immune complement system to substantially reduce the pathophysiological manifestations of CD55 deficiency in humans.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The microbiome sequencing data is linked to the National Center for Biotechnology Information (NCBI) BioProject ID PRJNA629392. The raw data, including the adat file used to generate proteomic analyses, the data analysis codes used for microbiome analyses and all other data that support the findings of this study, are available from the corresponding authors upon request to the extent allowed by all laws and institutional policies regarding confidentiality of patient clinical information. The following datasets were used in the study: Peditools; string db; maxikraken2 (v_1903_140GB) database; gnomAD v.2.1.1; and 1000genome.
References
Waldmann, T. A., Steinfeld, J. L., Dutcher, T. F., Davidson, J. D. & Gordon, R. S. Jr. The role of the gastrointestinal system in ‘idiopathic hypoproteinemia’. Gastroenterology 41, 197–207 (1961).
Ozen, A. et al. CD55 deficiency, early-onset protein-losing enteropathy, and thrombosis. N. Engl. J. Med. 377, 52–61 (2017).
Kurolap, A. et al. Loss of CD55 in eculizumab-responsive protein-losing enteropathy. N. Engl. J. Med. 377, 87–89 (2017).
Ozen, A. CHAPLE syndrome uncovers the primary role of complement in a familial form of Waldmann’s disease. Immunol. Rev. 287, 20–32 (2019).
Mevorach, D. Clearance of dying cells and systemic lupus erythematosus: the role of C1q and the complement system. Apoptosis 15, 1114–1123 (2010).
Holers, V. M. Complement and its receptors: new insights into human disease. Annu Rev. Immunol. 32, 433–459 (2014).
Kwan, W. H., van der Touw, W. & Heeger, P. S. Complement regulation of T cell immunity. Immunol. Res 54, 247–253 (2012).
Elvington, M., Liszewski, M. K. & Atkinson, J. P. Evolution of the complement system: from defense of the single cell to guardian of the intravascular space. Immunol. Rev. 274, 9–15 (2016).
Zipfel, P. F. & Skerka, C. Complement regulators and inhibitory proteins. Nat. Rev. Immunol. 9, 729–740 (2009).
Rother, R. P., Rollins, S. A., Mojcik, C. F., Brodsky, R. A. & Bell, L. Discovery and development of the complement inhibitor eculizumab for the treatment of paroxysmal nocturnal hemoglobinuria. Nat. Biotechnol. 25, 1256–1264 (2007).
Wong, E. K., Goodship, T. H. & Kavanagh, D. Complement therapy in atypical haemolytic uraemic syndrome (aHUS). Mol. Immunol. 56, 199–212 (2013).
Harris, C. L., Pouw, R. B., Kavanagh, D., Sun, R. & Ricklin, D. Developments in anti-complement therapy; from disease to clinical trial. Mol. Immunol. 102, 89–119 (2018).
Kurolap, A. et al. Eculizumab is safe and effective as a long-term treatment for protein-losing enteropathy due to CD55 deficiency. J. Pediatr. Gastroenterol. Nutr. 68, 325–333 (2019).
Di Narzo, A. F. et al. High-throughput identification of the plasma proteomic signature of inflammatory bowel disease. J. Crohns Colitis 13, 462–471 (2019).
Pickard, J. M., Zeng, M. Y., Caruso, R. & Núñez, G. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 279, 70–89 (2017).
Schwiertz, A. et al. Microbiota in Pediatric Inflammatory Bowel Disease. J. Pediatr. 157, 240–244.e1 (2010).
Giles, J. L., Choy, E., van den Berg, C., Morgan, B. P. & Harris, C. L. Functional analysis of a complement polymorphism (rs17611) associated with rheumatoid arthritis. J. Immunol. 194, 3029–3034 (2015).
Hillebrandt, S. et al. Complement factor 5 is a quantitative trait gene that modifies liver fibrogenesis in mice and humans. Nat. Genet. 37, 835–843 (2005).
Schatz-Jakobsen, J. A. et al. Structural basis for eculizumab-mediated inhibition of the complement terminal pathway. J. Immunol. 197, 337–344 (2016).
Fukuzawa, T. et al. Long lasting neutralization of C5 by SKY59, a novel recycling antibody, is a potential therapy for complement-mediated diseases. Sci. Rep. 7, 1080 (2017).
Zelek, W. M., Taylor, P. R. & Morgan, B. P. Development and characterization of novel anti-C5 monoclonal antibodies capable of inhibiting complement in multiple species. Immunology 157, 283–295 (2019).
Ardissino, G. et al. Complement functional tests for monitoring eculizumab treatment in patients with atypical hemolytic uremic syndrome: an update. Pediatr. Nephrol. 33, 457–461 (2018).
Lintner, K. E. et al. Early components of the complement classical activation pathway in human systemic autoimmune diseases. Front Immunol. 7, 36 (2016).
Yang, Y. et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am. J. Hum. Genet. 80, 1037–1054 (2007).
Saxena, K. et al. Great genotypic and phenotypic diversities associated with copy-number variations of complement C4 and RP-C4-CYP21-TNX (RCCX) modules: a comparison of Asian-Indian and European American populations. Mol. Immunol. 46, 1289–1303 (2009).
Dahm, A. E. A. et al. Elevated complement C3 and C4 levels are associated with postnatal pregnancy-related venous thrombosis. Thromb. Haemost. 119, 1481–1488 (2019).
Savelli, S. L. et al. Opposite profiles of complement in antiphospholipid syndrome (APS) and systemic lupus erythematosus (SLE) among patients with antiphospholipid antibodies (aPL). Front Immunol. 10, 885 (2019).
Medof, M. E., Walter, E. I., Rutgers, J. L., Knowles, D. M. & Nussenzweig, V. Identification of the complement decay-accelerating factor (DAF) on epithelium and glandular cells and in body fluids. J. Exp. Med. 165, 848–864 (1987).
Kostic, A. D. et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 22, 292–298 (2012).
Arthur, J. C. et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012).
Lloyd-Price, J. et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662 (2019).
Vujkovic-Cvijin, I. et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci. Transl. Med. 5, 193ra191 (2013).
Olm, M. R. et al. Necrotizing enterocolitis is preceded by increased gut bacterial replication, Klebsiella, and fimbriae-encoding bacteria. Sci. Adv. 5, eaax5727 (2019).
Shin, N. R., Whon, T. W. & Bae, J. W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 33, 496–503 (2015).
Blanton, L. V. et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 351, aad3311 (2016).
Heeschen, C. et al. Erythropoietin is a potent physiologic stimulus for endothelial progenitor cell mobilization. Blood 102, 1340–1346 (2003).
Ribatti, D. et al. Human erythropoietin induces a pro-angiogenic phenotype in cultured endothelial cells and stimulates neovascularization in vivo. Blood 93, 2627–2636 (1999).
Homburger, F. & Petermann, M. L. Studies on hypoproteinemia; familial idiopathic dysproteinemia. Blood 4, 1085–1108 (1949).
Parfitt, A. M. Familial neonatal hypoproteinaemia with exudative enteropathy and intestinal lymphangiectasis. Arch. Dis. Child 41, 54–62 (1966).
Srinivasan, M. & Roeske, R. W. Immunomodulatory peptides from IgSF proteins: a review. Curr. Protein Pept. Sci. 6, 185–196 (2005).
Sauter, R. J. et al. Functional relevance of the anaphylatoxin receptor C3aR for platelet function and arterial thrombus formation marks an intersection point between innate immunity and thrombosis. Circulation 138, 1720–1735 (2018).
Notaro, R. & Sica, M. C3-mediated extravascular hemolysis in PNH on eculizumab: mechanism and clinical implications. Semin Hematol. 55, 130–135 (2018).
Brucker, W. J. et al. An emerging role for endothelial barrier support therapy for congenital disorders of glycosylation. J. Inherit. Metab. Dis. 43, 880–890 (2020).
Ding, H., Kharboutli, M., Saxena, R. & Wu, T. Insulin-like growth factor binding protein-2 as a novel biomarker for disease activity and renal pathology changes in lupus nephritis. Clin. Exp. Immunol. 184, 11–18 (2016).
Azar, W. J. et al. IGFBP-2 enhances VEGF gene promoter activity and consequent promotion of angiogenesis by neuroblastoma cells. Endocrinology 152, 3332–3342 (2011).
Ozen, A., Comrie, W. A. & Lenardo, M. J. CD55 deficiency and protein-losing enteropathy. N. Engl. J. Med. 377, 1499–1500 (2017).
Mason, J. C., Lidington, E. A., Ahmad, S. R. & Haskard, D. O. bFGF and VEGF synergistically enhance endothelial cytoprotection via decay-accelerating factor induction. Am. J. Physiol. Cell Physiol. 282, C578–C587 (2002).
Mason, J. C. et al. Induction of decay-accelerating factor by cytokines or the membrane-attack complex protects vascular endothelial cells against complement deposition. Blood 94, 1673–1682 (1999).
Park, S. M. et al. Mapping the distinctive populations of lymphatic endothelial cells in different zones of human lymph nodes. PLoS ONE 9, e94781 (2014).
Zaneveld, J. R., McMinds, R. & Vega Thurber, R. Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2, 17121 (2017).
Eser, E. et al. The psychometric properties of the new Turkish generic health-related quality of life questionnaire for children (Kid-KINDL). Turk Psikiyatri Derg 19, 409–417 (2008).
Jaskowski, T. D., Martins, T. B., Litwin, C. M. & Hill, H. R. Comparison of three different methods for measuring classical pathway complement activity. Clin. Diagn. Lab. Immunol. 6, 137–139 (1999).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput Biol. 19, 455–477 (2012).
Wood, D. E., Lu, J. & Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 20, 257 (2019).
Ye, Y. & Doak, T. G. A parsimony approach to biological pathway reconstruction/inference for genomes and metagenomes. PLoS Comput. Biol. 5, e1000465 (2009).
Kanehisa, M., Sato, Y., Furumichi, M., Morishima, K. & Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 47, D590–D595 (2019).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 11, 119 (2010).
Kuznetsova, A., Brockhoff, P. & Christensen, R. lmerTest Package: tests in linear mixed effects models. J. Stat. Softw. https://doi.org/10.18637/jss.v082.i13 (2017).
Gold, L. et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 5, e15004 (2010).
Candia, J. et al. Assessment of variability in the SOMAscan assay. Sci. Rep. 7, 14248 (2017).
Cheung, F. et al. Web tool for navigating and plotting SomaLogic ADAT files. J. Open Res Softw. 5, 20 (2017).
Noris, M. et al. Dynamics of complement activation in aHUS and how to monitor eculizumab therapy. Blood 124, 1715–1726 (2014).
Emilsson, V. et al. Co-regulatory networks of human serum proteins link genetics to disease. Science 361, 769–773 (2018).
Eiseman, J. L. et al. Evaluation of plasma insulin-like growth factor binding protein 2 and Her-2 extracellular domain as biomarkers for 17-allylamino-17-demethoxygeldanamycin treatment of adult patients with advanced solid tumors. Clin. Cancer Res 13, 2121–2127 (2007).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Shapovalov, M. V. & Dunbrack, R. L. Jr. A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure 19, 844–858 (2011).
Tezcan, I., Berkel, A. I., Ersoy, F. & Sanal, O. Sağlıklı Türk çocukları ve erişkinlerde turbidometrik yöntemle bakılan serum immunoglobulin düzeyleri. Çocuk Sağlığı ve Hastalıkları Derg. 32, 649–656 (1996).
Slenter, D. N. et al. WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 46, D661–D667 (2017).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Szklarczyk, D. et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Acknowledgements
This work was supported in part by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH, BCBB Support Services Contract HHSN316201300006W/HHSN27200002, and The Marmara University, Scientific Research Projects Committee (BAPKO, grant no. SAG-C-TUP-230119-0018). We thank the Turkish National Society of Allergy and Clinical Immunology (TNSACI) for supporting travel expenses for the screening studies. We thank C. Kemper of the National Heart, Lung and Blood Institute for thoughtful editing of the final manuscript. We thank M. Quiñones, the Center for Human Immunology and the NIAID Microbiome Program, NIAID, NIH, for research support. We also thank A. Kiykim for patient care, A. Dalga and I. Tatli for technical assistance and H. Su and X. He for advice and assistance; D. Comrie, S. Kubo and J. Ravell for critical reading of the manuscript; and R. Kissinger for artwork. We thank important colleagues at Regeneron: A. N. Thomas for sample processing; C. Huang for biomarker analysis; H. Qiu, and E. Sook Yen for eculizumab analysis; and C. H. Lai, L. DeStefano and K. Donohue for total C5 analysis. Molecular graphics and analyses were performed with UCSF ChimeraX, developed by the Resource for Biocomputing, Visualization and Informatics at the University of California, San Francisco, with support from NIH R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, NIAID.
Author information
Authors and Affiliations
Contributions
A.O. and M.J.L. designed the trial and supervised all activities. A.O., M.J.L., I.V.-C., R. Apps, F.C., M.-D.W. and P.S. analyzed and interpreted data. O.H., J.T. and Y.B. supervised activities and interpreted data. R. Apps, G.F., B.S., Y.G.N., K.D.-N. and G.A. conducted experiments. Y.Z., A.L.S., C.L., C.A. and C.L.D. performed genetic sequencing and data analyses. A.O., S.B., E.K.-A., S.B.E., N.K., M.S., E. Tutar, D.E., B.A., S.S., F.O., D.K.U., A.I., O.B.S., G.K., E. Topal, E.S., R.H.J.H., S.N.G., A.B., I.O. and B.D. provided patient care and collected data. D.E., E. Tutar, R. Artan and B.A. conducted gastrointestinal interventions. C.C. made histopathology evaluations. R.E. provided radiological assessment. A.O., S.D.C. and M.J.L. wrote the paper. All authors reviewed, edited and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors M.J.L. and A.O. have a pending patent on C5 inhibitor treatment of CHAPLE. B.S. is a former SomaLogic, Inc. (Boulder, CO, USA) employee and a company shareholder. O.H., Y.G.N., M.-D.W. and K.D.-N. are employees of Regeneron Pharmaceuticals Inc., a biopharma company. There are no conflicts of interest to report for the remaining coauthors.
Additional information
Peer review information Nature Immunology thanks Kevin Marchbank, Stuart Tangye and Chack-Yung Yu for their contribution to the peer review of this work. Peer reviewer reports are available. L. A. Dempsey was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Demographic, clinical and laboratory characteristics of the patients with CHAPLE disease enrolled in the study.
Reference ranges are indicated in parenthesis. NA: Not assessed, NP: New patient, WBC: White blood count, ANC: Absolute neutrophil count, ALC: Absolute lymphocyte count lymphocyte count, Hgb: Hemoglobin, Plt: Platelet count. † CD55 NCBI Reference Sequence: NM_000574. * the lowest values in the past (more than 1 year before therapy) are indicated in the ‘past’ columns. ** the highest values in the past (more than 1 year before therapy) are indicated in the ‘past’ columns. The serum immunoglobulins compared to age-matched serum immunoglobulin reference ranges of Turkish children (Ref. 67). Peri-Tx: peri-treatment, referring to the past 12 months before the start of eculizumab until the end of the observation period.
Extended Data Fig. 2 Clinical symptoms, laboratory findings and growth percentiles of CHAPLE patients prior to and on eculizumab treatment.
Past: In the lifetime of the patient. Peri-Tx: Peri-treatment, referring to the past 12 months before the start of eculizumab until the end of the observation period. Current refers to the most recent measurement or the observation period when a particular patient has been on regular eculizumab Tx. + present, - absent, N/A: Not assessed. * We detected a recanalization of the narrowed segment in the thrombotic vessel in P5 during the follow up exam on eculizumab. However, there was no documented recovery of any of the thromboembolic complications in other patients during the indicated periods of follow up. ** P12 had ischemic gliotic foci in brain imaging. As identified by appropriate imaging including abdominal ultrasonography (USG) and/or doppler USG, and/or vascular imaging using a computed tomography or magnetic resonance imaging. § Growth percentiles not presented in certain columns; P2 was an adult during the peri-Tx period, and data not available in P14 and P15 for respective periods. † Abdominal pain cannot be assessed in P13 due to verbal immaturity and corresponding assessment was made for discomfort and abnormal crying behavior.
Extended Data Fig. 3 Medical interventions received by CHAPLE patients prior to and during eculizumab treatment.
Past: In the lifetime of the patient. Peri-Tx: Peri-treatment, referring to the past 12 months before the start of eculizumab until the end of the observation period. Current refers to the most recent measurement or the presence of a finding during the observation period when a particular patient has been on regular eculizumab Tx. *A repeat bowel surgery had been planned in P8 prior to eculizumab. Likewise, P3 had been planned to undergo bowel surgery before eculizumab. P15 underwent 2 separate bowel resection surgeries and received 39 days of intensive care support during the postoperative period, including mechanical ventilation. ** ‘Other invasive intervention’ includes placement of a central access device in P8 to deliver frequent albumin transfusions; intracardiac blood clot removal surgery, chest tube placement to drain pneumothorax and a cranial surgery to alleviate brain hemorrhage in P4; a percutaneous endoscopic gastrostomy port to support enteral nutrition in P12; a transjugular intrahepatic portosystemic shunt (TIPS) to relieve portal hypertension in P16. † P8 received intravenous (IV) antibiotics for sepsis and metronidazole for Giardia lamblia enteritis in the past. P4 and P12 experienced cardiovascular problems during the induction period of eculizumab, leading to dose skipping or termination of treatment, respectively. Eculizumab Tx was restarted in P12 after a period off- therapy, with no recurrence of similar manifestations. Fecal assessment during diarrheal episodes revealed Cryptosporidium parvum in P1 and P13 prior to eculizumab, who were treated with nitazoxanide uneventfully. The column for past medication before 1 year prior to eculizumab is blank for P13 due to young age and onset of symptoms during the past year. § Respiratory support refers to mechanical ventilation and/or oxygen treatment. PLE: Protein-losing enteropathy.
Extended Data Fig. 4 The effect of dose spacing between injections on albumin, trough eculizumab concentration, and complement markers during the maintenance therapy.
Blood samples were obtained prior to each injection. The horizontal line on left y axis shows lower range for albumin. Calculated mean ± S.D. values for dose intervals during the indicated time spans, or between two consecutive injections are presented in days. ECMb: eculizumab.
Extended Data Fig. 5 Variable dose spacing or interrupted therapy.
a, Normal serum protein levels sustained when switching from biweekly doses to monthly eculizumab injections in P1. Timing of eculizumab doses in relation to serum albumin and total protein concentrations for P1 in top panel, and immunoglobulin isotypes IgG, IgM and IgG in the bottom panel. b. Timeline plotting the incidence and severity of the indicated symptoms in P4 in relation to timing of eculizumab injection at various dosing intervals. Serum albumin levels plotted at the top of the figure with the horizontal bar showing normal range. Each arrow above the x-axis shows an eculizumab injection with the accompanying numbers illustrating days from the previous dose. The top line shows inability to feed (F) ranging from normal appetite (white) to intake completely abandoned (dark purple). The second line shows the presence (purple) or absence (white) of facial edema (E) in purple whereas the third line shows stool formation (S) on a scale from firm (white) to watery (dark gray). The fourth line quantifies the number of bowel movements (M) from 0-1 (white) to >11 (black). The fifth and sixth lines show the incidences of vomiting (V) from none (white) to >3 times per day (dark blue) and severity of nausea (N) from none (white) to severe (green), respectively. The last line shows severity of abdominal pain (P), ranging from none (white) to severe (dark brown). c. Serum albumin, total protein (T. protein), immunoglobulin G (IgG), and vitamin B12 concentrations for P3 before and after treatment at t = 0. Horizontal yellow bars for indicated parameters show normal range. Vertical dashed lines show eculizumab injections and purple bars dose skipping periods with the indicated numbers showing intervals between two consecutive injections.
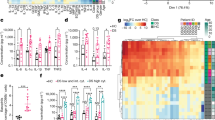
Extended Data Fig. 6 Biological significance of proteomic changes in serum.
a, Top-ranking pathways (Wikipathways) plotted against normalized enrichment score (Ref. 68,69). Note, the enrichment of the pathways did not reach adjusted statistical significance level of false discovery rate (FDR) < 0.05. b, Enrichment plots from pre-ranked analysis of log2 fold changes (FC) in protein abundances of patients with respect to age-matched controls, showing rank-based ordering of proteins belonging to immunoglobulin Pfam domain PF13927 at baseline. Similar plots are generated for indicated Post-treatment (Post-Tx) timepoints using log2 FC in protein abundances calculated with respect to patient baseline values. PF13927, found to be enriched in the STRING analysis (Ref. 70), was added to the Wikipathways gene set to compute the normalized enrichment score (NES) and FDR.
Supplementary information
Supplementary Information
Supplementary Figs. 1–16, Tables 1–2 and text.
Rights and permissions
About this article
Cite this article
Ozen, A., Kasap, N., Vujkovic-Cvijin, I. et al. Broadly effective metabolic and immune recovery with C5 inhibition in CHAPLE disease. Nat Immunol 22, 128–139 (2021). https://doi.org/10.1038/s41590-020-00830-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-020-00830-z
This article is cited by
-
Pozelimab: First Approval
Drugs (2023)
-
CD55-deficiency in Jews of Bukharan descent is caused by the Cromer blood type Dr(a−) variant
Human Genetics (2023)
-
Beyond IBD: the genetics of other early-onset diarrhoeal disorders
Human Genetics (2023)
-
Special issue: the genetics of early onset inflammatory bowel disease (IBD) and diarrheal disorders
Human Genetics (2023)
-
Precision medicine in monogenic inflammatory bowel disease: proposed mIBD REPORT standards
Nature Reviews Gastroenterology & Hepatology (2023)