Abstract
Metabolic reprograming toward aerobic glycolysis is a pivotal mechanism shaping immune responses. Here we show that deficiency in NF-κB-inducing kinase (NIK) impairs glycolysis induction, rendering CD8+ effector T cells hypofunctional in the tumor microenvironment. Conversely, ectopic expression of NIK promotes CD8+ T cell metabolism and effector function, thereby profoundly enhancing antitumor immunity and improving the efficacy of T cell adoptive therapy. NIK regulates T cell metabolism via a NF-κB-independent mechanism that involves stabilization of hexokinase 2 (HK2), a rate-limiting enzyme of the glycolytic pathway. NIK prevents autophagic degradation of HK2 through controlling cellular reactive oxygen species levels, which in turn involves modulation of glucose-6-phosphate dehydrogenase (G6PD), an enzyme that mediates production of the antioxidant NADPH. We show that the G6PD–NADPH redox system is important for HK2 stability and metabolism in activated T cells. These findings establish NIK as a pivotal regulator of T cell metabolism and highlight a post-translational mechanism of metabolic regulation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The RNA-seq datasets have been deposited in the Gene Expression Omnibus under accession no. GSE155576. The mass spectrometry data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with dataset identifier PXD020943 and https://doi.org/10.6019/PXD020943. Other datasets generated during the current study are available from the corresponding author upon reasonable request. Source data are provided with this paper.
Change history
12 February 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41590-021-00892-7
References
Durgeau, A., Virk, Y., Corgnac, S. & Mami-Chouaib, F. Recent advances in targeting CD8 T-cell immunity for more effective cancer immunotherapy. Front. Immunol. 9, 14 (2018).
Geltink, R. I. K., Kyle, R. L. & Pearce, E. L. Unraveling the complex interplay between T cell metabolism and function. Annu. Rev. Immunol. 36, 461–488 (2018).
Lim, A. R., Rathmell, W. K. & Rathmell, J. C. The tumor microenvironment as a metabolic barrier to effector T cells and immunotherapy. Elife 9, e55185 (2020).
Almeida, L., Lochner, M., Berod, L. & Sparwasser, T. Metabolic pathways in T cell activation and lineage differentiation. Semin. Immunol. 28, 514–524 (2016).
Chang, C.-H. et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251 (2013).
Buck, M. D., Sowell, R. T., Kaech, S. M. & Pearce, E. L. Metabolic instruction of immunity. Cell 169, 570–586 (2017).
Rivadeneira, D. B. & Delgoffe, G. M. Antitumor T-cell reconditioning: improving metabolic fitness for optimal cancer immunotherapy. Clin. Cancer Res. 24, 2473–2481 (2018).
Li, X. et al. Navigating metabolic pathways to enhance antitumour immunity and immunotherapy. Nat. Rev. Clin. Oncol. 16, 425–441 (2019).
Hashimoto, M. et al. CD8 T cell exhaustion in chronic infection and cancer: opportunities for interventions. Annu. Rev. Med. 69, 301–318 (2018).
McLane, L. M., Abdel-Hakeem, M. S. & Wherry, E. J. CD8 T cell exhaustion during chronic viral infection and cancer. Annu. Rev. Immunol. 37, 457–495 (2019).
Patsoukis, N. et al. PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat. Commun. 6, 6692 (2015).
Rashida Gnanaprakasam, J. N., Wu, R. & Wang, R. Metabolic reprogramming in modulating T cell reactive oxygen species generation and antioxidant capacity. Front. Immunol. 9, 1075 (2018).
Ronquist, G. & Theodorsson, E. Inherited, non-spherocytic haemolysis due to deficiency of glucose-6-phosphate dehydrogenase. Scand. J. Clin. Lab. Invest. 67, 105–111 (2007).
Stanton, R. C. Glucose-6-phosphate dehydrogenase, NADPH, and cell survival. IUBMB Life 64, 362–369 (2012).
Ho, H.-Y., Cheng, M.-L. & Chiu, D. T. Glucose-6-phosphate dehydrogenase—from oxidative stress to cellular functions and degenerative diseases. Redox Rep. 12, 109–118 (2007).
Yang, H.-C. et al. The redox role of G6PD in cell growth, cell death, and cancer. Cells 8, 1055 (2019).
Yang, K. & Chi, H. mTOR and metabolic pathways in T cell quiescence and functional activation. Semin. Immunol. 24, 421–428 (2012).
Maciver, N. J., Michalek, R. D. & Rathmell, J. C. Metabolic regulation of T lymphocytes. Annu. Rev. Immunol. 31, 259–283 (2013).
Saravia, J., Raynor, J. L., Chapman, N. M., Lim, S. A. & Chi, H. Signaling networks in immunometabolism. Cell Res. 30, 328–342 (2020).
Shyer, J. A., Flavell, R. A. & Bailis, W. Metabolic signaling in T cells. Cell Res. 30, 649–659 (2020).
Sun, S.-C. The non-canonical NF-κB pathway in immunity and inflammation. Nat. Rev. Immunol. 17, 545–558 (2017).
Dejardin, E. The alternative NF-κB pathway from biochemistry to biology: pitfalls and promises for future drug development. Biochem. Pharmacol. 72, 1161–1179 (2006).
Li, Y. et al. Cell intrinsic role of NF-κB-inducing kinase in regulating T cell-mediated immune and autoimmune responses. Sci. Rep. 6, 22115 (2016).
Wang, J., Saffold, S., Cao, X., Krauss, J. & Chen, W. Eliciting T cell immunity against poorly immunogenic tumors by immunization with dendritic cell-tumor fusion vaccines. J. Immunol. 161, 5516–5524 (1998).
Ahn, E. et al. Role of PD-1 during effector CD8 T cell differentiation. Proc. Natl Acad. Sci. USA 115, 4749–4754 (2018).
Sasaki, Y. et al. NIK overexpression amplifies, whereas ablation of its TRAF3-binding domain replaces BAFF:BAFF-R-mediated survival signals in B cells. Proc. Natl Acad. Sci. USA 105, 10883–10888 (2008).
Overwijk, W. W. et al. Tumor regression and autoimmunity after reversal of a functionally tolerant state of self-reactive CD8+ T cells. J. Exp. Med. 198, 569–580 (2003).
Pearce, E. L., Poffenberger, M. C., Chang, C.-H. & Jones, R. G. Fueling immunity: insights into metabolism and lymphocyte function. Science 342, 1242454 (2013).
Mizushima, N. Autophagy: process and function. Genes Dev. 21, 2861–2873 (2007).
Kaur, J. & Debnath, J. Autophagy at the crossroads of catabolism and anabolism. Nat. Rev. Mol. Cell Biol. 16, 461–472 (2015).
Lamark, T., Svenning, S. & Johansen, T. Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem. 61, 609–624 (2017).
Mehta, M. M. et al. Hexokinase 2 is dispensable for T cell-dependent immunity. Cancer Metab. 6, 10 (2018).
Murray, S. E. et al. NF-κB–inducing kinase plays an essential T cell–intrinsic role in graft-versus-host disease and lethal autoimmunity in mice. J. Clin. Invest. 121, 4775–4786 (2011).
Bollyky, P. L. et al. CD44 costimulation promotes FoxP3+ regulatory T cell persistence and function via production of IL-2, IL-10, and TGF-β. J. Immunol. 183, 2232–2241 (2009).
Tucker, E. et al. A novel mutation in the Nfkb2 gene generates an NF-κB2 ‘super repressor’. J. Immunol. 179, 7514–7522 (2007).
Yu, J. et al. T cell-intrinsic function of the noncanonical NF-κB pathway in the regulation of GM-CSF expression and experimental autoimmune encephalomyelitis pathogenesis. J. Immunol. 193, 422–430 (2014).
Filomeni, G., De Zio, D. & Cecconi, F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 22, 377–388 (2015).
Kamat, J. P. & Devasagayam, T. P. Nicotinamide (vitamin B3) as an effective antioxidant against oxidative damage in rat brain mitochondria. Redox Rep. 4, 179–184 (1999).
Choi, H. J., Jang, S.-Y. & Hwang, E. S. High-dose nicotinamide suppresses ROS generation and augments population expansion during CD8+ T cell activation. Mol. Cells 38, 918–924 (2015).
Pretsch, W., Charles, D. J. & Merkle, S. X-linked glucose-6-phosphate dehydrogenase deficiency in Mus musculus. Biochem. Genet. 26, 89–103 (1988).
Ham, M. et al. Glucose-6-phosphate dehydrogenase deficiency improves insulin resistance with reduced adipose tissue inflammation in obesity. Diabetes 65, 2624–2638 (2016).
Pauken, K. E. et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165 (2016).
Wei, S. C. et al. Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. Cell 170, 1120–1133.e17 (2017).
Varanasi, S. K., Jaggi, U., Hay, N. & Rouse, B. T. Hexokinase II may be dispensable for CD4 T cell responses against a virus infection. PLoS ONE 13, e0191533 (2018).
Brightbill, H. D. et al. Conditional deletion of NF-κB-inducing kinase (NIK) in adult mice disrupts mature B cell survival and activation. J. Immunol. 195, 953–964 (2015).
Gershon, T. R. et al. Hexokinase-2-mediated aerobic glycolysis is integral to cerebellar neurogenesis and pathogenesis of medulloblastoma. Cancer Metab. 1, 2 (2013).
Zhou, X. et al. The deubiquitinase Otub1 controls the activation of CD8+ T cells and NK cells by regulating IL-15-mediated priming. Nat. Immunol. 20, 879–889 (2019).
Xiao, G., Harhaj, E. W. & Sun, S. C. NF-κB-inducing kinase regulates the processing of NF-κB2 p100. Mol. Cell 7, 401–409 (2001).
Ouyang, C. et al. Transforming growth factor (TGF)-β-activated kinase 1 (TAK1) activation requires phosphorylation of serine 412 by protein kinase A catalytic subunit α (PKACα) and X-linked protein kinase (PRKX). J. Biol. Chem. 289, 24226–24237 (2014).
Acknowledgements
We thank Genentech for providing the Map3k14 flox mice. This study was supported by a grant from the National Institutes of Health (NIH) (no. GM84459 to S.-C.S.) and partially supported by a grant from the National Research Foundation funded by the South Korean government (no. NRF-2020R1A3B2078617 to J.B.K.). We thank the flow cytometry, sequencing and microarray, proteomics and animal facility of the shared resources at the MD Anderson Cancer Center, supported by the NIH/National Cancer Institute Cancer Center Support Grant no. P30CA016672. The Proteomics Facility is also supported by NIH grant no. IS10OD012304 and the Cancer Prevention and Research Institute of Texas grant no. RP130397.
Author information
Authors and Affiliations
Contributions
M.G. designed and performed the research, prepared the figures and wrote part of the manuscript. X. Zhou designed and performed the research and prepared the figures. L.Z., Z.J., J.-Y.Y., X.X., J.Y., Y.S. and X.C. contributed to the experiments. X. Zheng and J.W. performed the RNA-seq data analysis. J.H.S. and J.B.K. contributed the G6pdmut mouse materials. H.D.B. contributed the Map3k14 flox mice, and S.-C.S. supervised the work and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
J.Y. is an employee of Precision for Medicine. H.D.B. is an employee of Genentech. The other authors declare no competing interests.
Additional information
Peer review information Peer reviewer reports are available. Nature Immunology thanks Stephen Ley and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Zoltan Fehervari was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 NIK regulates T cell exhaustion and antitumor immunity.
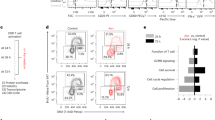
a, genotyping PCR of R26StopFLMap3k14 (Map3k14Tg), Map3k14+/+CreER, and R26StopFLMap3k14-CreER (Map3k14TgCreER) mice, showing the PCR products of NIK-GFP in Map3k14Tg allele and CreER. b, Immunoblot analysis of NIK expression in tamoxifen-treated Map3k14+/+CreER (WT) and Map3k14TgCreER (iTg) mice. c, Schematic of experimental design for producing B16F10 tumor-bearing NIKiTg and WT control mice. Each mouse was injected s.c. with 5 × 105 B16F10 cells (WT = 9, iTg=7). d,e, Flow cytometric analysis of the frequency and absolute cell number of CD4 and CD8 T cells in the tumor (d) or draining lymph node (e) of day 18 B16F10 tumor-implanted NIK-iTg and WT mice (d, WT: n = 4; iTg: n = 5; e, n = 6 per genotype). f,g, Flow cytometric analysis of the frequency and absolute number of CD44+CXCR3+ CD8 effector T cells in the draining lymph node (f) or tumor (g) of day 18 B16F10 tumor-implanted NIK-iTg and wildtype mice (f, n = 6 per genotype; g, n = 4 per genotype). h,i, Flow cytometric analysis of the frequency and absolute number of PD1+Tim3+ CD8+ T cells (h) or IFNγ-producing PD1+Tim3+ CD8 T cells (i) in the tumor of day 18 B16F10-implanted NIKiTg and wildtype control mice (h,i, n = 4 per genotype). Data are representative of three independent experiments. Summary data are shown as mean ± s.e.m. with P values determined by two-tailed Student’s t test.
Extended Data Fig. 2 Ectopic expression of NIK improves CD8 T cell function in adoptive T cell therapy.
a,b, Flow cytometric analysis of the frequency of total donor CD8 T cells (a) and IFNγ-producing donor CD8 T cells (b) in the tumor of B16F10 melanoma-bearing B6.SJL recipient mice adoptively transferred with wildtype or NIKiTg Pmel1 CD8 T cells as described in Fig. 2o (n = 4 per genotype). c, In vitro cytotoxicity assay of wildtype (WT) and NIKiTg Pmel1 CD8 effector T cells towards B16F10 tumor cells at the indicated effector to tumor (E:T) ratios. d-f, Schematic of experimental design (d) and flow cytometric analysis of apoptosis based on caspase 3 cleavage (e) or proliferation based on CFSE dilution (f) of tumor-infiltrating wildtype (WT) and NIKiTg OT-I CD8 T cells in B16-OVA-tumor bearing B6.SJL mice adoptively transferred with in vitro activated and CFSE-labeled WT or NIKiTg CD8 T cells for 7 days (e, n = 3 per genotype). Data are representative of two independent experiments. Summary data are shown as mean ± s.e.m. with P values determined by two-tailed Student’s t test.
Extended Data Fig. 3 NIK deficiency has no effect on mRNA expression of glycolysis-regulatory genes and glucose uptake but promotes lysosomal localization of HK2.
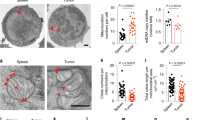
a, Seahorse analysis of OCR under baseline (no treatment) and maximum or stressed (injection of FCCP) conditions in untreated Map3k14tKO (tKO) or wildtype (WT) naïve CD8 T cells. Data are shown as a representative plot (upper) and summary graphs (lower, each circle represents a well). b, Volcano plot of RNA sequencing analysis of differentially expressed genes in Map3k14tKO OT-I CD8 T cells relative to WT OT-I CD8 T cells, activated with anti-CD3 plus anti-CD28 for 24 hr. c, Immunoblot analysis of the indicated proteins in WT or Map3k14tKO OT-I CD8 T cells stimulated with anti-CD3 plus anti-CD28 for the indicated time periods. d,e, Flow cytometric analysis of glucose uptake using in vitro activated (d) or B16F10 tumor-infiltrating (e) CD8 T cells. f, Flow cytometry analysis of HK2 expression in tumor-infiltrating CD8 T cells of B16F10-bearing wildtype (WT) or Hk2tKO mice, showing the specificity of the HK2 staining. g, Immunoblot analysis of HK2 and the indicated control proteins in the cytoplasmic (Cy) and lysosomal (Ly) fractions of wildtype (WT) and Map3k14tKO CD8 T cells. h, Immunoblot analysis of the indicated proteins in whole cell lysates of wildtype or Map3k14tKO CD8 T cells treated for the indicated time points in the presence (+) or absence (–) of lysosomal inhibitors, E64D plus pepstatin A. i, Summary of h based on densitometric quantification of three independent experiments. Data are representative of two (b,f) or three(a,c-e,g-h) independent experiments. Summary data are shown as mean ± s.e.m. with P values determined by two-tailed Student’s t test.
Extended Data Fig. 4 T cell-specific deletion of HK2 impairs T cell metabolism and immune responses against tumorigenesis and bacterial infection.
a,b, Genotyping PCR (a) and immunoblot (b) analysis of the Hk2tKO (tKO) and wildtype (WT) control mice. c,d, Seahorse analysis of basal ECAR (measured after glucose injection, Glc) and maximum ECAR (measured after oligomycin injection, Oligo) (c) and Seahorse analysis of baseline OCR (no treatment) and maximum OCR (FCCP injection) (d) in Hk2tKO or wildtype OT-I CD8 T cells either naïve or activated with anti-CD3 and anti-CD28 for 24 h. e, Flow cytometric analysis of the expression level of PD1 and Tim3 in CD8 T cells isolated from day-20 tumor of the B16F10-implanted Hk2tKO and wildtype control mice (n = 4 per genotype). f,g, Flow cytometric analysis of the frequency and absolute number of IFNγ-producing CD8 effector T cells in splenocytes (f) and bacterial burden in the spleen (g, presented as a representative image and a summary graph based on multiple mice) of Hk2tKO or wildtype mice infected i.v. with L. monocytogenes (1 × 105 CFU/mouse) for 7 (f) or 4 (g) days (f, n = 4 per genotype; g, n = 6 per genotype). h, Flow cytometric analysis of IFNγ-producing CD8 T cells in the spleen of the indicated mouse strains infected for 84 days with LCMV clone 13(4 × 106 PFU/mouse), restimulated in vitro with 3 μg/ml LCMV gp33-41 peptide for 14 h with monensin added during the last hour; WT(n = 6), Map3k14tKO(n = 6) and Hk2tKO(n = 3). Data are representative of two (f-h) or three (a-e) independent experiments. Summary data are shown as mean ± s.e.m. with P values determined by two-tailed Student’s t test.
Extended Data Fig. 5 NIK-mediated stimulation of antitumor T cell responses requires HK2.
Flow cytometric analysis of the frequency and absolute cell number of CD4 and CD8 T cells (a), PD1+Tim3+ CD8 T cells (b) and flow cytometric analysis of the expression level (MFI) of PD1 and Tim3 in CD8 T cells (c) in day-27 tumor of wildtype (WT), NIKiTg (iTg), NIKiTgHk2iKO (iTg-iKO) mice injected s.c. with 5 × 105/mouse MC38 colon cancer cells (a,b, WT: n = 4; iTg: n = 4; iTg-iKO: n = 3). Data are representative of two independent experiments. Summary data are shown as mean ± s.e.m. with P values were determined by two-tailed Student’s t test.
Extended Data Fig. 6 Noncanonical NF-κB activation is not required for HK2 stabilization or T cell metabolism.
a, Immunoblot analysis of HK2 expression and p100 processing (as a measure of noncanonical NF-kB activation) in WT or Nfkb2Lym1/+ (Lym1/+) OT-I CD8 T cells stimulated with anti-CD3 plus anti-CD28 for the indicated time periods. b,c, Seahorse analysis of basal ECAR (after glucose injection) and maximum ECAR (after oligomycin injection) (b) and Seahorse analysis of baseline OCR (no treatment) and maximum OCR (FCCP injection) (c) of WT or Nfkb2Lym1/+ (Lym1/+) OT-I CD8 T cells, either naïve or activated with anti-CD3 and anti-CD28 for 24 h, under baseline and stressed conditions. d, Coimmunoprecipitation analysis of NIK-HK2 interaction using whole-cell lysates of 293 cells transfected with (+) or without (–) the indicated expression vectors. Data are representative of three independent experiments. Summary data are shown as mean ± s.e.m. based on multiple wells (each circle represents a well) with P values determined by two-tailed Student’s t test.
Extended Data Fig. 7 ROS is involved in HK2 degradation in NIK-deficient CD8 T cells.
a, Flow cytometric analysis of ROS levels in Map3k14tKO OT-I CD8 T cells activated with anti-CD3 plus anti-CD28 for 48 h in the presence of NAM or solvent control DMSO. b,c, Immunoblot analysis of HK2 expression in Map3k14tKO (tKO) or wildtype (WT) OT-I CD8 T cells activated with anti-CD3 plus anti-CD28 for 48 h in the presence of the antioxidant N-acetylcysteine (NAC) or medium control (b) or the indicated ROS inducers and DMSO control (c). d, Immunoblot analysis of HK2 in wildtype (WT) or Atg5-tKO CD8 T cells activated with anti-CD3 plus anti-CD28 for 48 h in the presence of the indicated ROS inducers or medium control. e, Immunoblot analysis of HK2 and the indicated loading controls in lysosomal (Ly) and cytoplasmic (Cy) fractions of wildtype (WT) and Map3k14tKO CD8 T cells activated with anti-CD3 plus anti-CD28 for 48 h in the presence of NAC or medium control. f, Flow cytometry analysis of the mass and membrane potential of mitochondria in wildtype (WT) and Map3k14tKO (tKO) CD8 T cells stimulated with anti-CD3 plus anti-CD28 for 24 h. g, Ratio of reduced (GSH) and oxidized (GSSG) forms of glutathione in Map3k14tKO (upper), NIKiTg (lower), or wildtype (WT) control OT-I CD8 T cells activated in vitro with anti-CD3 plus anti-CD28 for 48 h. Data are representative of three independent experiments. Summary data are shown as mean ± s.e.m. with P values determined by two-tailed Student’s t test.
Extended Data Fig. 8 G6PD phosphorylation by NIK and role in regulating HK2 expression and T cell metabolism.
a, Seahorse analysis of basal ECAR (measured after glucose injection, Glc) and maximum ECAR (measured after oligomycin injection, Oligo) and Seahorse analysis of baseline OCR (no treatment) and maximum OCR (FCCP injection) in G6PDmut or wildtype (WT) control T cells activated by anti-CD3 plus anti-CD28 for 48 h. b,c, Immunoblot analysis of the indicated proteins (b) and ROS detection (c) in wildtype or Nfkb2lym1/+ (Lym1/+) CD8 T cells, which were either not treated (NT) or stimulated with anti-CD3 plus anti-CD28 for the indicated time periods. d, CoIP analysis of NIK-G6PD interaction (upper) and immunoblot analysis of HA-NIK and Flag-G6PD expression in whole-cell lysates (WCL) of 293 cells transfected with (+) or without (–) Flag-G6PD and HA-NIK. e, Phos-tag SDS PAGE and Immunoblot analysis of phosphorylated (p-) and total G6PD as well as GST-NIK in an in vitro kinase assay mix containing 500 ng recombinant His-G6PD and the indicated amounts of recombinant GST-NIK. f, NIK-induced G6PD phosphorylation sites identified by mass spectrometry analysis of G6PD phosphorylated in vitro by NIK. g, G6PD activity in 293 T cells transiently transfected with Flag-tagged wildtype G6PD or the indicated G6PD mutants along with either an empty vector or HA-NIK. h,i, Schematic of experimental design (h) and immunoblot analysis of G6PD expression in T cells isolated from bone marrow chimeric mice constructed using G6PDmut bone marrow cells reconstituted with either an empty vector (Vector) or the indicated G6PD expression vectors (i). For a and g, data are shown as representative plots, each circle represents a well. Data are representative of one (f) or three (a-e,g-i) independent experiments. Summary data are shown as mean ± s.e.m. with P values determined by two-tailed Student’s t test.
Supplementary information
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 7
Unprocessed western blots.
Source Data Fig. 8
Statistical source data.
Source Data Fig. 8
Unprocessed western blots.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 7
Unprocessed western blots.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 8
Unprocessed western blots.
Rights and permissions
About this article
Cite this article
Gu, M., Zhou, X., Sohn, J.H. et al. NF-κB-inducing kinase maintains T cell metabolic fitness in antitumor immunity. Nat Immunol 22, 193–204 (2021). https://doi.org/10.1038/s41590-020-00829-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-020-00829-6
This article is cited by
-
NF-κB in biology and targeted therapy: new insights and translational implications
Signal Transduction and Targeted Therapy (2024)
-
Cellular metabolism regulates the differentiation and function of T-cell subsets
Cellular & Molecular Immunology (2024)
-
The effects of metabolism on the immune microenvironment in colorectal cancer
Cell Death Discovery (2024)
-
Aberrant accumulation of NIK promotes tumor growth by dysregulating translation and post-translational modifications in breast cancer
Cancer Cell International (2023)
-
Hexokinase 2 confers radio-resistance in hepatocellular carcinoma by promoting autophagy-dependent degradation of AIMP2
Cell Death & Disease (2023)