Abstract
The antimicrobial functions of neutrophils are facilitated by a defensive armamentarium of proteins stored in granules, and by the formation of neutrophil extracellular traps (NETs). However, the toxic nature of these structures poses a threat to highly vascularized tissues, such as the lungs. Here, we identified a cell-intrinsic program that modified the neutrophil proteome in the circulation and caused the progressive loss of granule content and reduction of the NET-forming capacity. This program was driven by the receptor CXCR2 and by regulators of circadian cycles. As a consequence, lungs were protected from inflammatory injury at times of day or in mouse mutants in which granule content was low. Changes in the proteome, granule content and NET formation also occurred in human neutrophils, and correlated with the incidence and severity of respiratory distress in pneumonia patients. Our findings unveil a ‘disarming’ strategy of neutrophils that depletes protein stores to reduce the magnitude of inflammation.
Similar content being viewed by others
Main
Neutrophils are endowed with highly specialized features to combat infections. They accumulate receptors, proteases, antimicrobial peptides and proteins inside cytoplasmic granules that mediate the production of reactive oxygen species or DNA-based extracellular traps (NETs), both of which are highly toxic and allow for effective containment of pathogens1,2. Several types of granules coexist in the neutrophil cytoplasm3, including primary (azurophilic) granules that store myeloperoxidase (MPO), secondary (specific) and tertiary (gelatinase) granules and secretory vesicles, each of which carry distinct types of cargo1,4. The different granules form at different stages of granulopoiesis in the bone marrow5,6, and the synthesis of granule proteins declines as neutrophils differentiate7,8. In contrast, receptors needed for sensing, migration and phagocytosis are mostly synthesized once they reach the circulation7,8.
After only 6–10 h in the circulation9, neutrophils rapidly disappear from blood10,11. However, this does not imply immediate elimination, as they can be found within the vessels or parenchyma of multiple organs, with a preference for the spleen, bone marrow and lungs10. The homeostatic infiltration of granule-laden neutrophils, a process enabled by a cell-intrinsic program that relies on the chemokine receptor CXCR2 and the core clock protein Bmal1 (ref. 12), can be potentially damaging to highly vascularized tissues such as the lungs. Indeed, lungs are highly susceptible to neutrophil-mediated damage, which can cause respiratory dysfunction and death, as shown in murine models of lung injury13,14,15. In humans, neutrophil-mediated acute vascular damage followed by pulmonary edema and distress is common in patients with underlying pneumococcal infections or other pre-existing conditions13,16.
Here, we investigated whether specific mechanisms prevent neutrophil-inflicted toxicity in tissues. We found that the neutrophil proteome spontaneously ‘disarms’ as the neutrophils lose granule content during their lifetime in circulation. This resulted in a blunted capacity to produce NETs, rendering the neutrophils less toxic before they reached tissues, and protected the lungs from injury both in mice and in patients with pneumonia.
Results
The neutrophil proteome changes during the day
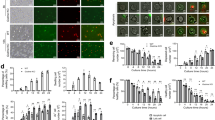
Neutrophils undergo transcriptional changes while in the circulation12. However, because transcriptional changes take a relatively long time (in the range of hours) and diurnal transcriptional programs in mouse neutrophils favor their migration into tissues, we examined alternative mechanisms that might protect the tissues from the toxic activity of neutrophils. We undertook a proteomic approach to explore the changes in protein content in neutrophils at different times after their release from the bone marrow. Because the number of neutrophils that can be purified from the blood of naïve mice at night (when neutrophils are released from the bone marrow) or at noon (when neutrophils have spent several hours in the circulation)17 is low, we increased the number of night-like neutrophils released from the bone marrow by a one-time intraperitoneal injection at zeitgeber time 4 (ZT4, or 4 h after lights on) of AMD3100, an antagonist for CXCR4 that induces the rapid mobilization of neutrophils from the bone marrow18. To increase the number of day-like neutrophils in circulation we injected blocking antibodies against endothelial selectins (1 mg kg−1, intravenously at ZT5 at days −5, −3 and −1 before collection)12,17 (Extended Data Fig. 1a). The proteomic analysis of these night- and day-like neutrophils isolated at ZT5 from the blood of wild-type mice yielded 6,677 proteins (Supplementary Table 1). A total of 171 proteins were differentially detected when comparing the night-like versus the day-like group, with proteins involved in regulation of the cytoskeleton (Arghdib, Rufy3), adhesion (Thbs1, Itgam), inflammasome (Nlrp3, Nlrp4e) and vesicular transport (Vps33b) (Fig. 1a) enriched in night-like neutrophils compared to day-like neutrophils. The differential expression of identified proteins in night- versus day-like neutrophils, such as VEGFR1, CD74, CD16/32, CD63 or CD14, was confirmed by flow cytometry (Extended Data Fig. 1b). Gene ontology analyses (Supplementary Table 2) indicated involvement of the differentially expressed proteins in immune defense, cytokine signaling and angiogenesis (Extended Data Fig. 1c). Night-like neutrophils were enriched for pathways linked to c-Kit signaling and platelet production, possibly reminiscent of their time in the bone marrow, as well as GTPase Rho signaling (Fig. 1b), consistent with the enhanced migratory capacity of ‘younger’ neutrophils12. Day-like neutrophils showed upregulation of protein synthesis, pathogen sensing and respiration (Fig. 1b), suggesting specialization for effector functions and enhanced TLR signaling, as previously suggested12,19. Comparison of the neutrophil proteome with transcriptional datasets associated with diurnal time12 showed poor correlation (Extended Data Fig. 1d), suggesting that the proteomic changes are not driven by direct transcriptional regulation. These results therefore identify prominent changes in the neutrophil proteome that are associated with the time spent in the circulation.
a, Volcano plot of the total proteome showing proteins with FDR < 0.01 (calculated as stated in the Methods from single samples of 60 million neutrophils pooled from nine mice (night) and six mice (day)). A positive Z score represents an increase in nighttime (fresh) over daytime (aged) neutrophils. Color represents the functional category for each protein (top right). b, Gene-set enrichment analysis of the proteomics data showing pathways with P < 0.15 enriched in aged (red) or fresh (blue) neutrophils. The size of the bubble-plot represents overlap between the query and the gene set (bottom right). Vertical lines indicate queries with multiple pathways. c, Volcano plot of the proteomic dataset showing granule proteins with a Z score >2. Colors show the granule type for each protein (top right). d, Z stack maximum projection of neutrophils stained for primary granules with MPO and counterstained with DAPI. Scale bar, 5 μm. e, Quantification of neutrophil granule contents during a full diurnal cycle. Curves are repeated (dashed line) to better appreciate the circadian pattern. Dark phase is shown in gray; n = 30 cells per time point. f, Neutrophil elastase (NE) activity in plasma. Curves are repeated (dashed line) to better appreciate the circadian pattern. Dark phase is shown in gray; n = 4 mice per time point. Data in e and f are shown as mean ± s.e.m., and circadian oscillation P values were determined by amplitude versus zero two-tailed t-test analysis (see Methods).
Circulating neutrophils degranulate in the steady state
Based on the proteomic data, proteins stored in the various types of granules (azurophilic, specific and tertiary/gelatinase granules) were enriched in night-like neutrophils compared to day-like neutrophils (Fig. 1c and Supplementary Table 3). Enrichment in granule proteins was also detected in the proteome of circulating neutrophils from AMD3100-mobilized mice compared to daytime neutrophils (Extended Data Fig. 1e,f and Supplementary Table 4). Further analysis in the reactome database indicated that proteins differentially detected in the night- versus day-like neutrophils were associated with degranulation (Extended Data Fig. 2a), altogether suggesting a progressive loss of granules as neutrophils circulated.
Cytometric analysis of wild-type circulating neutrophils purified every 4 h (ZT1, ZT5, ZT9 (daytime); ZT13, ZT17, ZT21 (nighttime)) revealed circadian oscillations of side-scatter properties (Extended Data Fig. 2b), a feature that reflects cell granularity. To assess if these variations in side scatter were caused by degranulation, we stained cytospins of Ly6G+ blood neutrophils purified at the same times as for MPO, a protein of azurophilic granules, and analyzed granule content by high-resolution confocal microscopy (Fig. 1d). The maximal abundance of MPO+ granules was observed at ZT17 (midnight, when neutrophils leave the marrow), while the valley was at ZT1 (Fig. 1e and Extended Data Fig. 2c), indicating progressive loss of primary granules. MPO+ granule content in neutrophils was in antiphase with elastase activity in plasma (Fig. 1f), suggesting the release of granule content into blood. A 12-h shift in the animal light cycle for three weeks completely readjusted the pattern of granule content in neutrophils (Extended Data Fig. 2d), indicating that degranulation was circadian in nature. Finally, Ly6G+ neutrophils from lungs, spleen and liver had a lower MPO+ granule content compared to Ly6G+ neutrophils in the blood (Extended Data Fig. 2e,f). Thus, neutrophils undergo homeostatic degranulation in the circulation before they enter tissues.
Neutrophils progressively lose NET-forming capacity
Proteases stored in the azurophilic granules degrade the nuclear histones and promote chromatin decondensation during NET formation20 and granule proteins have been identified in the NET proteome21. Among the proteins found in the proteomic analyses, those associated with NETs (Supplementary table 3) were enriched in the proteome of night-like neutrophils (Fig. 2a), suggesting that NET release is more prevalent at this time. To directly examine whether the formation of NETs varied during the course of a day, we treated Ly6G+ neutrophils purified at ZT5 or ZT13 from the blood of wild-type mice with phorbol myristate acetate (PMA), a NET-inducing compound, and analyzed for the presence of extracellular DNA decorated with MPO protein and citrullinated histone 3 (cit-H3). We found that neutrophils at ZT13 (night) formed more NETs than at ZT5 (daytime) (Fig. 2b), revealing a correlation between diurnal azurophilic granule content and NET-forming capacity in blood Ly6G+ neutrophils. Next, we assessed the presence of NETs by whole-mount immunostaining with the same markers in cremaster muscles of wild-type and Lyz2CreMcl1fl/fl mice, which are neutropenic22, after ischemia-reperfusion. Many more NETs were detectable in the muscles of mice at ZT13 than at ZT5, while no NETs were detected in the Lyz2CreMcl1fl/fl mice (Fig. 2c). Thus, proteomic changes and degranulation of circulating neutrophils temporally coincide with loss of NET-forming ability.
a, Volcano plot of the neutrophil proteome showing proteins found in NETs (red dots), and enrichment of these proteins in nighttime neutrophils. b, Ex vivo NET-formation assay. Neutrophils sorted at ZT13 (nighttime) or ZT5 (daytime) were stimulated with PMA or vehicle to induce NETs and were stained for cit-H3 and DNA (left). Colocalization of both markers was used to quantify NET formation, as shown in the bar graph (right); n = 3 mice per time point. c, Representative images (left) and quantification (right) of in vivo NET formation in the cremaster muscle subjected to ischemia/reperfusion at nighttime (ZT13) or at daytime (ZT5). Triple colocalization of MPO, DNA and cit-H3 was used to define and quantify the area of NETs (red; arrowheads). Neutropenic Mcl1∆N mice (Lyz2Cre;Mcl1fl/fl mice) were used as controls and showed no NETs. Dotted areas are shown magnified in the bottom panels; n = 3 mice per condition. Scale bars, 50 μm. Bars show mean ± s.e.m. *P < 0.05; NS, not significant, as determined by unpaired two-tailed t-test analysis.
Proteome changes are driven by CXCR2 and Bmal1
Diurnal changes in neutrophil transcription and migration are controlled by a cell-intrinsic mechanism, in which the expression of the chemokine Cxcl2 is regulated by the molecular clock protein Bmal1 and leads to the cell-autonomous, diurnal activation of neutrophils by signaling through CXCR2 (ref. 12). We therefore tested whether this mechanism caused the proteome changes. Treatment with CXCL2 induced degranulation in isolated wild-type Ly6G+, circulating neutrophils (Fig. 3a). Analysis of the granule content of blood neutrophils from MRP8CRE Cxcr2fl/fl (referred to as CXCR2ΔN hereafter)12, which have a neutrophil-specific deficiency in CXCR2, at ZT5 and ZT13, showed no differences in MPO+ granule content or in side-scatter properties in blood neutrophils, compared with neutrophils from wild-type mice (Fig. 3b and Extended Data Fig. 2b). CXCR2ΔN neutrophils degranulated in response to PMA and lipopolysaccharide (LPS) (Extended Data Fig. 3a,b), indicating that degranulation could be induced in these cells. We also found a loss of diurnal oscillations in granule content in blood neutrophils from Cxcl2−/− mice (Fig. 3b). Neutrophils from CXCR2ΔN and Cxcl2−/− mice also showed no differences in NET formation between ZT5 and ZT13 (Fig. 3c). In chimeric mice transplanted with equal mixes of bone marrow from DsRed+ wild-type and Cxcl2−/− mice, Cxcl2−/− neutrophils had a higher granule content than wild-type neutrophils analyzed at the same time (ZT5; Fig. 3d), suggesting that CXCL2 signaled in neutrophils in an autocrine fashion to control granule content.
a, Ex vivo stimulation of sorted neutrophils with CXCL2 induces degranulation (right), as quantified by confocal imaging of MPO-stained neutrophils (left); n = 29 (−) and 32 (+) cells. Scale bar, 2 μm. b, Diurnal quantification of granule content in neutrophils from WT, CXCR2∆N or Cxcl2−/− mice, showing a loss of diurnal fluctuation in CXCR2-deficient (n = 38 cells at ZT5 and 40 cells at ZT13) or CXCL2-deficient neutrophils (n = 31 cells at ZT5 and 14 cells at ZT13), compared to their WT counterparts (n = 30 cells at ZT5 and 34 cells at ZT13). c, Ex vivo NET-formation assays with sorted neutrophils stimulated with PMA or vehicle control, at nighttime (ZT13) or daytime (ZT5). NETs were quantified by triple colocalization of cit-H3, DNA and MPO in confocal micrographs (left; scale bar, 25 μm). Each mouse was normalized to its vehicle control and NET formation at ZT13 and ZT5 is compared (right). CXCL2-deficient (n = 3 mice per time) and CXCR2-deficient (n = 6 mice at ZT5 and 2 mice at ZT13) neutrophils showed loss of diurnal fluctuation in NET formation compared with WT cells (n = 6 mice at ZT5 and 4 mice at ZT13). d, CXCL2 signaling causes cell-autonomous degranulation, as shown by analysis of MPO+ granules in neutrophils from bone marrow chimeras reconstituted with DsRed+ wild-type and DsRedNEG Cxcl2−/− donors; scale bar, 5 μm; n = 3 mice. Bars show mean ± s.e.m. **P < 0.01; NS, not significant, as determined by paired (a,d) or unpaired (b,c) two-tailed t-test analysis.
Circadian expression of Cxcl2 in neutrophils is controlled by the transcription factor Bmal1 (ref. 12). MRP8CRE Arntlfl/fl mice, which have a neutrophil-specific deletion of Arntl (which encodes Bmal1, referred hereafter as Bmal1ΔN) showed no circadian differences in MPO+ granule content between ZT5 and ZT13 (Extended Data Fig. 4a) and NET formation (Extended Data Fig. 4b) in blood Ly6G+ neutrophils compared to neutrophils from wild-type controls, suggesting that Bmal1 controlled the changes in the neutrophil proteome. Proteome analysis in Ly6G+ neutrophils purified at ZT5 (day) or ZT13 (night) from the blood of Bmal1ΔN mice (Extended Data Fig. 4c and Supplementary Table 5) indicated that Bmal1ΔN neutrophils did not show circadian changes in granule proteins or in NET-associated proteins (Extended Data Fig. 4d,e). These data indicated that Bmal1 and signaling through CXCL2–CXCR2 controlled the changes in granule content and the loss in the capacity to form NETs in neutrophils.
The severity of lung injury varies during the day
To test whether the diurnal changes in the neutrophil proteome influenced the outcome of inflammatory responses in tissues, we used a model of endotoxin and antibody-induced acute lung inflammation (ALI)15,23, which is dependent on neutrophils and NETs24,25,26, and recapitulates the pulmonary injury observed in transfused patients27. We used ALI-prone Balb/c mice, which displayed normal diurnal variations in neutrophil number and phenotype (Extended Data Fig. 5a–c)12. Lungs from Balb/c mice treated to induce ALI (by intraperitoneal injection of 0.1 mg kg−1 LPS 24 h before intravenous injection of 0.5 mg kg−1 of an anti-H2d antibody) showed evidence of NETs, defined by staining for MPO, DNA and cit-H3 (Fig. 4a and Supplementary Videos 1 and 2)24,26. To assess the kinetics of NET formation in vivo during ALI, we performed high-speed multichannel intravital imaging of the lung microcirculation28. Neutrophils were identified by Ly6G staining, while NET-like structures where identified by the rapid extrusion of Sytox-green-labeled DNA from Ly6G+ neutrophils, as a result of intravenous injection of the dye 5 min before imaging (Fig. 4b and Supplementary Video 3). The Sytox-green+ extrusions were inhibited by the PAD4 inhibitor chloramidine injected intravenously 1 h before induction of ALI (Fig. 4c), indicating that they represented NETs. Continuous tracking of neutrophil behavior for 40 min showed higher release of Sytox-green+ extrusions at ZT13 compared with ZT5 (Fig. 4c and Supplementary Video 3). The enhanced NET production at night was detectable minutes after induction of ALI and was sustained for the rest of the experiment (Fig. 4c). In contrast, the number of Ly6G+ neutrophils was similar at ZT5 and ZT13 in untreated and in chloramidine-treated mice (Extended Data Fig. 6a–c). Because platelets have been involved in neutrophil activation and NET formation during ALI15,25, we assessed their possible involvement in the observed differences between ZT5 and ZT13. We found a sharp increase in the number of platelets in the pulmonary vessels in mice with ALI; however, their numbers were comparable at ZT5 and ZT13 (Extended Data Fig. 6b), suggesting that the number of neutrophils or platelets did not influence the diurnal changes in pulmonary NET formation.
a, Presence of NETs in lungs. Maximum projections of confocal Z stack series of cleared lungs from control mice (basal) or mice with antibody-induced ALI are shown. Lungs were stained against cit-H3, MPO and DNA, and some NETs are shown (arrowheads). Scale bar, 30 μm. Representative images of n = 3 cleared lungs per condition. b, Time series of intravital imaging captures of NET-like structures in the lungs of ALI-induced mice. NET-like structures were defined as free DNA extruded out of Ly6G+ neutrophils. Scale bar, 10 μm. See also Supplementary Video 1. c, Quantification of NET-like structures as shown in b, and normalized to the number of neutrophils in mice in which ALI was performed at nighttime (ZT13, blue line) or daytime (ZT5, red line), or in mice treated with chloramidine (at ZT5, dashed gray line). Time course (left panel) and area under the curve values (right panel); in both, n = 15 fields from four mice per condition. Individual data points are not shown here as this graph uses a mean ± s.e.m. value for the area under the curve calculated from the data shown in the left panel. d, Representative images of longitudinal CT series of edema formation at 0 or 21 min after inducing ALI ZT13 or ZT5. Note the increased edema (red) at night. Background bone signal (gray) is shown for anatomical positioning. e, Quantification of the images in d. Volume of edema was increased at ZT13 (blue; n = 7 mice) relative to ZT5 (red; n = 7 mice). Mice treated with chloramidine are shown as a control (gray, n = 4 mice). f, Survival of mice subjected to ALI at ZT13 (blue, n = 18 mice) or ZT5 (red, n = 21 mice), or treated with chloramidine (gray, n = 11 mice). Data are shown as mean ± s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001, as determined by one-way ANOVA with Dunnet’s multiple comparison test (c), two-way ANOVA (e) or log-rank (two-tailed Mantel–Cox) test (f).
To evaluate the effects of the diurnal changes in NET formation in the inflamed lungs, we measured the kinetics of edema formation, which is caused by infiltration of plasma in the alveolar space. Intravenous injection with Evans blue dye after ALI induction at ZT5 indicated a marked increase in pulmonary vascular permeability during ALI, compared to LPS-only treated mice (Extended Data Fig. 7a), which was specific to the lungs. We then used computerized tomography (CT) to track the in vivo dynamics of edema in mice in which we induced ALI at ZT5 (day) or at ZT13 (night). We found a rapid and sharp increase in water content of the lungs, which occurred earlier and was higher in ZT13- compared with ZT5-induced mice (Fig. 4d,e). This correlated with reduced survival of mice in which ALI was induced at ZT13 compared to those induced at ZT5 (Fig. 4f). Finally, treatment with chloramidine 1 h before induction at ZT5 protected mice from edema and death (Fig. 4e,f), indicating that the release of NETs was a primary cause of ALI. Thus, the severity of lung injury during ALI displays diurnal patterns that correlate with the granule content and the capacity of neutrophils to form NETs.
Variations in the neutrophil proteome drive lung injury
To assess whether neutrophil-intrinsic variations in granule content and NET formation caused lung inflammation, we used Bmal1ΔN and MRP8CRE Cxcr4fl/fl mice, which lacked CXCR4, a negative regulator of CXCR2 signaling12, specifically in neutrophils (hereafter CXCR4ΔN). Neutrophils from Bmal1ΔN and CXCR4ΔN mice share transcriptional and migratory properties with night and day neutrophils, respectively12. MPO immunofluorescence and transmission electron microscopy (TEM) imaging of blood Ly6G+ neutrophils purified at ZT5 revealed higher azurophilic granule content in Bmal1ΔN neutrophils compared with wild-type neutrophils, and comparable with that of ZT13 wild-type neutrophils (Fig. 5a,b). In contrast, CXCR4ΔN neutrophils at ZT5 had strong reductions in azurophilic granule content compared with wild-type neutrophils at any time point (Fig. 5a,b). NET formation in response to PMA was elevated in ZT5 Bmal1ΔN neutrophils, and strongly reduced in CXCR4ΔN neutrophils, relative to ZT5 wild-type neutrophils (Fig. 5c). Thus, variations in granule content and NET formation are driven by a neutrophil-intrinsic program.
a, Confocal micrographs (left) and quantification (right) of primary granules in neutrophils from WT mice at night (ZT13, n = 33 cells from three mice) or daytime (ZT5, n = 30 cells from three mice), and from mutant mice (n = 50 cells from three CXCR4∆N mice and 41 cells from three Bmal1∆N mice, both at ZT5); scale bar, 2 μm. b, TEM images (left) and quantification of electrodense azurophilic granules (right) in WT and mutant mice (all at ZT5), showing increased granule content in Bmal1∆N and reduced in CXCR4∆N neutrophils compared with WT cells; n = 19 cells from three mice each; scale bar, 1 μm. c, Ex vivo NET formation in sorted WT, Bmal1∆N or CXCR4∆N neutrophils stimulated with PMA or vehicle as control; n = 3 mice per condition. d, Quantification of NET-like structures normalized to the number of neutrophils during ALI in Bmal1∆N (purple line) or CXCR4∆N (blue line) mice. Time course and elevations from baseline (gray area) are shown in the left panel, and the areas under the curve are shown in the right panel (individual data points are not shown here as this graph uses a mean ± s.e.m. value for the area under the curve calculated from the data shown in the left panel). Experiments were performed at ZT5. Data from wild-type mice at ZT5 (light gray, dashed) and ZT13 (dark gray, dashed) from Fig. 4c are shown for reference; n = 15 fields from three mice for each condition. e, Representative images of longitudinal CT series of edema (red) in mutant mice subject to ALI (left) at ZT5, and quantification of the edema volume over time (right); n = 7 mice per genotype. f, Survival curves for Bmal1∆N (n = 19 mice) and CXCR4∆N (n = 12 mice) mice subjected to ALI at ZT5. Data are shown as mean ± s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant, as determined by one-way ANOVA with Dunnet’s multiple comparison test (a–d), two-way ANOVA (e) or two-sided log-rank (Mantel–Cox) test (f).
To test whether the diurnal changes in the susceptibility to ALI were caused by changes in neutrophil granularity, we analyzed the responses of Bmal1ΔN and CXCR4ΔN mice to induction of ALI at ZT5. Intravital microscopy analyses showed faster and elevated NET formation in the lungs of Bmal1ΔN mice compared with lungs of CXCR4ΔN mice, which were similar to the differences found at ZT13 and ZT5, respectively, in wild-type mice (Fig. 5d). Intravital microscopy and flow cytometry showed similar numbers of neutrophils and platelets, as well as the number of interactions with each other, within the microvasculature of wild-type, Bmal1ΔN and CXCR4ΔN mice (Extended Data Fig. 6d–g), indicating an otherwise normal behavior of intravascular neutrophils in these mice. We found marked elevation in pulmonary edema and reduced survival in Bmal1ΔN mice during ALI induced at ZT5 compared with CXCR4ΔN mice (Fig. 5e,f and Supplementary Video 4). The protection from pathology correlated with preservation of vascular permeability in the lungs of CXCR4ΔN mice after intravenous injection of Evans blue (Extended Data Fig. 7b,c). Intravital imaging in wild-type mice allowed visualization of two distinct NET-like formation events, ‘flowing’ NETs, in which the extruded NETs were immediately washed away by the bloodstream, and ‘adherent’ NETs, in which the extruded DNA remained adherent to the vasculature (Extended Data Fig. 7d and Supplementary Video 5). The frequency of adherent NETs was elevated compared with flowing NETs in Bmal1ΔN mice, but not in wild-type or CXCR4ΔN mice (Extended Data Fig. 7e), suggesting that the ability of neutrophils to deposit NETs onto the endothelium enhanced vascular damage in Bmal1ΔN mice.
Next, we tested the causal relationship between the diurnal proteome changes in neutrophils and the diurnal susceptibility to inflammation in the Bmal1ΔN mice. Neutrophils from Bmal1ΔN mice had a constitutively high MPO+ granule content and NET-forming capacity (Extended Data Fig. 4a,b) and reduced survival during ALI at both ZT5 or ZT13 (Extended Data Fig. 8a). CXCR4ΔN neutrophils, in turn, had a similar loss in circadian granule content and NET formation (Extended Data Fig. 8b,c) and higher survival during ALI at both ZT5 or ZT13 (Extended Data Fig. 8a). Of note, the number of circulating neutrophils in Bmal1ΔN mice oscillated during the day, similar to wild-type mice (Extended Data Fig. 8d), whereas CXCR4ΔN mice display constitutively high neutrophil counts12, indicating that the granule content, but not the number of neutrophils, was associated with the severity of pulmonary inflammation. These observations indicated that impaired diurnal changes in the proteome and the granule content in neutrophils were related to the magnitude of inflammation.
Human neutrophils undergo proteome disarming
Clinical manifestations of inflammation display circadian oscillations in humans, and are associated with time-of-day variations in both the incidence and the severity of cardiovascular disease29,30,31. We tested whether neutrophil ‘disarming’ in humans could underlie the circadian changes in vascular inflammation. We measured neutrophil counts in blood and performed proteomic analysis, granule content and NET-formation assays in neutrophils isolated at 8:00, 14:00 and 17:00 (Extended Data Fig. 9a), when diurnal patterns in neutrophil number and phenotype are prominent in humans12, from ten healthy volunteers. Human neutrophils display a night-like phenotype (similar to murine ZT13) early in the morning, and progress toward a day-like phenotype (similar to murine ZT5) over the next 4–8 h (ref. 12). In our cohort, neutrophil counts increased over time and peaked at 14:00–19:00 (Fig. 6a), while the granularity of neutrophils, indicated by the side scatter by flow cytometry, decreased from 8:00 to 19:00 (Fig. 6b). Direct examination of granules in human neutrophils stained for MPO (Fig. 6c,d), as well as by TEM imaging (Fig. 6e), indicated a diurnal reduction in granule content, which peaked at 8:00 and showed a trough at 14:00.
a, Blood neutrophil counts in human volunteers at the different time points; n = 10 human volunteers per time point. b, Light-scattering (SSC-A) values for human neutrophils at different times, as measured by flow cytometry; n = 10 human volunteers. c, Quantification of primary granules from confocal images of human neutrophils from images in d; n = 150 cells from ten volunteers per time point. d, Representative images of granule content of human neutrophils, quantified in c. e, Transmission electron micrographs (left) of human neutrophils in the morning (8:00) and early afternoon (14:00), and quantification (right) showing reduced numbers of electrodense primary granules; n = 33 cells from three human volunteers per time point. f, Volcano plot of the human neutrophil proteome analyzed at 8:00 and 14:00, showing proteins with P < 0.001 (see Methods for human TMT proteomics). A negative Z score represents higher values at 8:00 over 14:00 neutrophils; n = 5 per time point. g, Volcano plot of granule proteins in the human neutrophil proteome, showing higher content in granule proteins (color-coded) at 8:00 compared with 14:00; n = 5 samples from healthy volunteers. Red dots and labels show differentially expressed proteins with P < 0.05, and black dots show all granule proteins. Label color indicates the granule type in which the protein is normally found (top right). h,i, Representative confocal images (h) and quantification (i) of ex vivo NET formation by human neutrophils stimulated with PMA or vehicle at the indicated time points; n = 10 volunteers per time point. j, ARDS severity shown as the pneumonia severity index (left) and intrahospital deaths (right) of patients entering the intensive care unit at different times of the day; n = 125 patients. Data are shown as mean ± s.e.m. **P < 0.01, as determined by unpaired two-tailed t-test analysis (f). P values for the circadian plots were calculated by the amplitude versus zero two-tailed t-test analysis (a–c,g,i). Maroon line in a–c, g and i shows the nonlinear COSINOR fit of the data.
Proteomic analyses of neutrophils isolated in density gradients at 8:00 and 14:00 from the blood of five healthy volunteers (Supplementary Table 6) indicated that, from a total of 1,918 proteins detected, around 9.7% were differentially enriched between 8:00 and 14:00 (P < 0.05; Fig. 6f). Gene ontology analyses revealed that the differentially detected proteins were in pathways related to vesicle-mediated transport, secretion, exocytosis or degranulation (Extended Data Fig. 9b and Supplementary Table 7), consistent with a loss of granule content in human neutrophils over time. Many of the individual proteins identified in the mouse analyses were not detected or did not show diurnal changes in the human samples (not shown), probably indicating a high variability among humans compared with co-housed and strain-matched mice. However, numerous granule proteins, such as MPO, MMP9 or LTF, were more abundant at 8:00 than at 14:00 in human neutrophils (Fig. 6g). Similar to mouse neutrophils, there was a poor correlation between transcript and protein amounts in human neutrophils (Extended Data Fig. 9c), suggesting that the proteomic changes were largely independent of transcriptional regulation. Ex vivo assays of human neutrophils purified using density gradients and treated with vehicle or PMA indicated a marked reduction in NET-forming capacity between 8:00 and 14:00 (Fig. 6h,i), corresponding with more NET-related proteins at 8:00 compared with 14:00 (Extended Data Fig. 9d). These observations suggested that diurnal degranulation also occurred in circulating human neutrophils.
Finally, we tested whether the changes in proteome, granule content and NET-forming activity in human neutrophils were associated with variations in the susceptibility to develop inflammation and/or its severity. We focused on an existing cohort of 5,334 patients with community-acquired pneumonia, who are at risk of developing acute respiratory distress syndrome (ARDS)32. Retrospective analysis of the human cohort indicated that, among the patients hospitalized with community-acquired pneumonia, 125 (2.3%) developed ARDS. Assessment of the temporal patterns of disease onset and severity (using the pneumonia severity index), by plotting the times of admission against the pneumonia index and death in this subset of patients, revealed consistent diurnal oscillations, with a peak around 9:00 and a trough at 17:00–19:00 for both parameters (Fig. 6j). These observations indicate that proteomic disarming occurs in human neutrophils and may influence the incidence and severity of inflammation (Extended Data Fig. 10 and Supplementary Video 6).
Discussion
Here, we describe the existence of a cell-intrinsic program in neutrophils that induced the progressive loss of granule proteins involved in inflammation and the formation of NETs, and provided a previously unrecognized layer of protection against the toxic activity of these leukocytes. The phenomenon of proteome disarming described here shares key features with the process of neutrophil ‘aging’12, in terms of reliance on CXCR2 signaling, regulation by Bmal1 and CXCR4 in a cell-intrinsic manner and similar temporal patterns (that is, peak at noon). This suggested that neutrophil aging and disarming might represent distinct manifestations of a core circadian program in neutrophils that allows anticipation of potential threats12, but also protects from excessive inflammation, vascular damage and death.
Beyond regulating the magnitude of inflammation, changes in the neutrophil proteome displayed temporal oscillations that aligned neutrophil activity with circadian cycles. The circadian transcriptional regulation of migration-associated genes by Bmal1 controls the migratory properties of neutrophils, and allows the infiltration of naïve tissues in anticipation of potential infections12. In addition to these transcription-mediated mechanisms, we uncovered here that the neutrophil proteome is also subject to circadian regulation, which blunts the ability of neutrophils to release toxic mediators and to produce NETs. This level of immune regulation may be particularly relevant for granulocytes, a group of innate immune cells that synthesize and store the majority of their early-response mediators inside dedicated organelles. Because the formation of granules and their content occurs as neutrophils differentiate in the bone marrow7,33, it is unlikely that changes of the granule proteome that occur in the circulation are transcriptionally regulated, as illustrated by the poor correlation between the proteome and the transcriptome in these cells. Our data, however, suggested a model in which upstream control of transcription of Cxcl2 by Bmal1 (ref. 12) allowed indirect regulation of the neutrophil proteome by the canonical circadian machinery. Conceptually, the temporal nature of this process in blood aligns with the evolutionary value of circadian oscillations in segregating mutually antagonistic processes, in this case potent antimicrobial responses and protection of the host against immune-mediated damage.
The circadian patterns in the granule content and elastase activity in plasma suggested the progressive discharge of granules while in the circulation, such that neutrophils that migrate into tissues during the day10 would have lost part of their antimicrobial arsenal, and would be, as such, less likely to elicit organ damage, as shown in the mouse model of ALI. Because neutrophil proteases are also important for the activation of many biological mediators, including cytokines, chemokines, growth factors and adhesion and pattern-recognition receptors34,35, and even modulate signaling cascades in other cells36,37, the programmed proteome changes reported here may have pleiotropic consequences in physiology. For instance, they predict that neutrophils that are newly released from the bone marrow in conditions of stress, such as obesity or cancer38,39, may be better suited to regulate physiological aspects of target organs than the aged neutrophils that enter the tissues at the end of their life cycle and have lost part of their cargo.
Our observations shed light into the particular susceptibility of the lungs to diurnal inflammation. Large numbers of neutrophils accumulate within the pulmonary microcirculation in mice, rabbits and nonhuman primates after stimulation or in the steady state10,18,40, and the lungs can provide a niche for effective antimicrobial responses41. The homeostatic accumulation of neutrophils in the lungs at the end of the light cycle in mice10 would predict higher susceptibility to inflammatory stimuli at this time. However, we found that neutrophils recovered from naïve lungs had reduced granularity, and that mutant mice with constitutive low levels of granules in neutrophils displayed permanent protection from ALI, suggesting a dominant contribution of the disarming process in tissue protection.
It has long been recognized that multiple inflammatory processes in humans display circadian periodicity42, and experimental models and retrospective studies have corroborated that not only the onset of inflammation, but also the severity of the inflammatory events manifest diurnal oscillations31. These events are often caused by neutrophil activation and by thrombosis, which in turn can be exacerbated in the presence of neutrophils or NETs15,25,43,44. Thus, our current findings in the context of ALI may extend to other inflammatory and thrombotic conditions affecting multiple organs, and suggest that interventions aimed at inducing controlled degranulation may effectively ‘disarm’ neutrophils and protect organs from catastrophic inflammation.
Methods
Mice
All experiments were performed in 7–18-week-old male C57BL/6 or Balb/c mice kept in a specific pathogen-free facility at Centro Nacional de Investigaciones Cardiovasculares (CNIC) under a 12 h light/12 h dark schedule (lights on at 7:00, off at 7:00), with water and chow available ad libitum. Bmal1∆N, CXCR4∆N and CXCR2∆N mutants in the C57BL/6 background have been described12. For the ALI experiments, Bmal1∆N and CXCR4∆N mice were back-crossed into the Balb/c background for at least six generations and we confirmed susceptibility to antibody-induced lung injury at this stage (not shown). All procedures with back-crossed mice were controlled using Cre-negative littermates. For bone marrow transplant experiments, WT donor bone marrow came from mice expressing DsRed under the control of the β-actin promoter (DsRedTg). No specific randomization method was followed in this study. All experimental procedures were approved by the Animal Care and Ethics Committee of CNIC and the regional authorities.
Proteomic analysis of fresh and aged mouse neutrophils with oxygen-18
Neutrophils were obtained from the blood of C57BL6 mice using different strategies. For night-like neutrophils, mice were treated with AMD3100 (2.5 mg kg−1 intraperitoneal) 1 h before collection. For day-like neutrophils, mice were treated with anti-P/E-selectin antibodies (1 mg kg−1 intravenous) for 5 d, every other day, before collection. Blood was obtained by collection in EDTA tubes to prevent coagulation. Red blood cells (RBCs) were precipitated using methylcellulose, and leukocytes in the top phase were collected and washed before purification by negative selection. Negative selection was performed by incubation with biotinylated antibodies against CD3e, TER119, B220, CD8, CD115, CD4, CD49b and CD117, then incubated with streptavidin-coated beads and applied in a magnet for negative selection of neutrophils. We obtained neutrophils with a purity over 84%. A total of 60 million neutrophils from each source were obtained for analysis.
After in-gel trypsin digestion, the ‘night-like’ samples were equally mixed with the ‘day-like’ samples, which were labeled with oxygen-18. The resulting tryptic peptide mixtures were subjected to nanoliquid chromatography coupled to mass spectrometry (MS) for protein identification. Peptides were injected onto a C-18 reversed-phase nanocolumn (75 μm internal diameter and 50-cm length, Acclaim PepMap100, Thermo Scientific) and analyzed in a continuous acetonitrile gradient consisting of 0–30% B in 240 min, 30–90% B in 3 min (B is 90% acetonitrile, 0.5% acetic acid). A flow rate of ~200 nl min−1 was used to elute peptides from the reversed-phase nanocolumn to an emitter nanospray needle for real-time ionization and peptide fragmentation on a Q Exactive mass spectrometer (Thermo Fisher). An enhanced Fourier transform-resolution spectrum (resolution was 70,000) followed by the tandem mass spectra (MS/MS) from the most intense 15 parent ions were analyzed along the chromatographic run (272 min). Dynamic exclusion was set at 30 s. For protein identification, MS/MS spectra were extracted and the charge state deconvoluted by Proteome Discoverer v.1.4.0.288 (Thermo Fisher). All MS/MS samples were analyzed using SEQUEST (Thermo Fisher) with a fragment ion mass tolerance of 20 ppm and a parent ion tolerance of 15 ppm. The carbamidomethyl of cysteine was specified in SEQUEST as a fixed modification. Oxidation of methionine was specified in SEQUEST as a variable modification. Peptide quantification from FullScan spectra and labeling efficiency calculation were performed as described using QuiXoT, a custom software written in C#. Statistical analysis was performed as reported45. The standardized variable Zq describes the log2 ratio for each protein corrected for the corresponding systematic error, in each experiment. Finally, data were plotted using R (v.3.6.1). The code will be provided on request. Statistical data on all proteomics analysis are presented in Supplementary Table 8.
Proteomic analysis of neutrophils by tandem mass tag
For proteomic analysis of human neutrophils, two million isolated cells were pelleted in 1.5-ml Eppendorf tubes and snap-frozen at −80 °C. On analysis of purity, the best five samples (96% mean purity) were used for multiplexed proteomic analysis at 8:00 and 14:00. For mouse neutrophils, 100,000 cells were sorted from Bmal1∆N mice at ZT5 or ZT13 (see Cytometry and cell sorting).
Total protein extracts from each sample were treated with 50 mM iodoacetamide and digested overnight at 37 °C with trypsin (Promega) at a 40:1 protein:trypsin (w/w) ratio using the Filter Aided Sample Preparation digestion kit (Expedeon) according to the manufacturer’s instructions. The resulting peptides were desalted with C-18 Oasis cartridges (Waters Corporation) using 50% acetonitrile (v/v) in 0.1% trifluoroacetic acid (v/v) as eluent, and vacuum-dried. The peptides were tandem mass tag (TMT)-labeled following the manufacturer’s instructions, desalted, separated into four fractions using the high pH reversed-phase peptide fractionation kit (Thermo Fisher) and analyzed using a Proxeon EASY nanoflow HPLC system (Thermo Fisher) coupled via a nanoelectrospray ion source (Thermo Fisher) to a Q Exactive HF Orbitrap mass spectrometer (Thermo Fisher). A C-18-based reverse phase separation was used with a 2-cm trap column and a 50-cm analytical column (EASY column, Thermo Fisher) in a continuous acetonitrile gradient consisting of 0–30% A for 240 min, 50–90% B for 3 min (A is 0.1% formic acid; B is 90% acetonitrile, 0.1% formic acid) at a flow rate of 200 nl min−1. Mass spectra were acquired in a data-dependent manner, with an automatic switch between MS and MS/MS using a top 15 method. MS spectra in the Orbitrap analyzer were in a mass range of 400–1,500 m/z and 120,000 resolution. Higher-energy collisional dissociation fragmentation was performed at 28 eV of normalized collision energy and MS/MS spectra were analyzed at 30,000 resolution in the Orbitrap.
All database searches were performed with Proteome Discoverer (v.2.1, Thermo Fisher) using SEQUEST-HT (Thermo Fisher) against a Uniprot database containing all sequences from human (accessed July 2018; 20,408 entries). For database searching, parameters were selected as follows: trypsin digestion with two maximum missed cleavage sites, precursor mass tolerance of 2 Da, fragment mass tolerance of 30 ppm. Methionine oxidation (+15.994915) was set as a variable modification. Lysine and peptide amino-terminal modification of +229.162932 Da, as well as cysteine carbamidomethylation of +57.021464 Da, were set as fixed modifications. The same MS/MS spectra collections were also searched against inverted databases constructed from the same target databases. Peptide identification from MS/MS data was performed using the probability ratio method. False discovery rates (FDR) of peptide identifications were calculated using the refined method. A 1% FDR was used as the peptide identification criterion. Each peptide was assigned only to the best protein proposed by the Proteome Discoverer algorithm.
Quantitative information from TMT reporter intensities was integrated from the spectrum level to the peptide level, and then to the protein level based on the WSPP model using the Generic Integration Algorithm (GIA)46,47. Briefly, for the human experiment, for each sample i, the values \(x_{qps} = {\mathrm{log}}_2A_i/C_i\) were calculated, where Ai is the intensity of the TMT reporter of the 14:00 sample from the individual i in the MS/MS spectrum, s, coming from peptide, p, and protein, q, and Ci is the intensity of the TMT reporter from the 8:00 sample from the same individual. Protein quantifications where further integrated among the five comparisons in the human experiment, obtaining the averaged value \(\left( {{x_q}^\prime } \right)\) with their corresponding weights \(\left( {{w_q}^\prime } \right)\) (ref. 47) for every protein.
In the mouse TMT experiments, for each sample i, the values xqps = log2Ai/Ci were calculated, where Ai is the intensity of the TMT reporter from the individual i in the MS/MS spectrum, s, coming from peptide, p, and protein, q, and Ci is the averaged intensity of the TMT reporters from all the samples in the given TMT experiment. The log2 ratio of each peptide (xqp) was calculated as the weighted mean of its spectra, the protein values (xq) were the weighted mean of its peptides and the grand mean (\(\bar x\)) was calculated as the weighted mean of all the protein values47. The statistical weights of spectra, peptides and proteins (wqps, wqp and wq, respectively) and the variances at each one of the three levels (\(\sigma _s^2\), \(\sigma _p^2\) and \(\sigma _q^2\), respectively) were calculated as described47. Protein quantifications where further integrated among the corresponding replicates from the same condition, obtaining averaged values \(\left( {{x_q}^\prime } \right)\) with their corresponding weights \(\left( {{w_q}^\prime } \right)\) (ref. 47) for every protein.
In all cases, protein abundance changes were then expressed in standardized units (Zq), from which statistical significance of the changes in terms of the P value were calculated. P values were corrected for multiple hypothesis testing by controlling for the FDR. Abundance changes of proteins in the compared samples were considered significant at 1% FDR.
Data were analyzed using R (v.3.6.1). The analysis code will be provided on request. Statistical data on all proteomics analysis is presented in Supplementary Table 8.
Proteomics–transcriptomics correlations
To analyze the correlation between proteomics and transcriptomics data for fresh and aged neutrophils, we compared our proteomics data with previously generated RNA-sequencing data on fresh (sorted at ZT13 from blood) and aged neutrophils (sorted at ZT5 from blood) available in Gene Expression Omnibus under accession number GSE86619 (ref. 12). To this end we obtained a set of common proteins and genes from both datasets, compared the proteomics Z score and the sequencing log2 FC (both inform about the relative enrichment in fresh neutrophils) and calculated the regression (Spearman) statistics using R (v.3.6.1). We performed similar analyses for paired transcriptome and proteome data from human samples.
Cytometry and cell sorting
Cytometric analyses were performed using a Sony SP6800 Spectral Analyzer (Sony Biotechnology). Analysis was performed using FlowJo v.X (Tree Star). Cell sorting experiments were performed using a FACS Aria cell sorter (BD Biosciences). All analyses were conducted at the Cellomics Unit of the CNIC. The following antibodies and streptavidin conjugates were used in this study:
Antibody | Clone | Species | Manufacturer |
|---|---|---|---|
CD16-APC | 3G8 | Human | BD |
CD62L-FITC | DREG56 | Human | BD |
CD11b-PE | M1/70 | Human/mouse | Tonbo Biosciences |
Ly6G-AF647 | 1A8 | Mouse | eBioscience |
CD45-APC/Cy7 | 104 | Mouse | BioLegend |
CXCR2-PerCP-Cy5.5 | SA044G4 | Mouse | BioLegend |
CXCR4-PE | 2B11 | Mouse | eBioscience |
CD41-PE | MWReg30 | Mouse | eBioscience |
CD62L-FITC | MEL-14 | Mouse | eBioscience |
CD11b-BV510 | M1/70 | Mouse | BioLegend |
CD31-APC | 390 | Mouse | eBioscience |
CD31 | 2H8 | Human/mouse | Thermo Fisher |
MPO-biotin | Polyclonal | Human/mouse | R&D (biotinylated in-house) |
cit-H3 (R2 + R8 + R17) | Polyclonal | Human/mouse | Abcam |
Goat-anti-hamster-AF405 | Polyclonal | Hamster | Jackson ImmunoResearch |
Goat-anti-rabbit-AF647 | Polyclonal | Rabbit | Thermo Fisher |
Streptavidin-AF488 | Not applicable | Mouse/human/rat | BioLegend |
In vivo NET formation during ischemia/reperfusion
The cremaster muscle was exteriorized as reported12, and ischemia was achieved by occlusion of the inflowing and outflowing vessels by clamping the tissue connecting the muscle and the animal’s body with a 15-mm Micro Serrefine clamp (Fine Science Tools) for 45 min. Reperfusion was achieved by removal of the clamp. At 15 min after reperfusion, mice were euthanized and cremaster muscle was removed for analysis. Excised muscles were fixed in 4% paraformaldehyde (PFA) at 4 °C overnight and then washed three times in PBS containing 0.5% Triton X-100 (PBST) and blocked for 2 h in PBST 25% fetal bovine serum (FBS) at room temperature with shaking. Staining was performed using anti-cit-H3, anti-MPO and anti-CD31 in 10% FBS-PBST overnight at 4 °C with shaking. Then, secondary antibodies were stained in 10% FBS-PBST for 4 h at room temperature. Samples were then washed and mounted in Mowiol 4-88 (Mw = 31,000; Sigma). Imaging of whole-mount cremaster muscles was performed using a Nikon A1R confocal system coupled to a Nikon Eclipse-Ti inverted microscope with the following lines: diode 402 nm, argon laser 457, 476, 488, 514 nm, diode 561 nm, HeNe laser 642 nm using a Plan Apo 10×/0.45 dry objective and the software NIS Elements AR v.4.30.02 (Build 1053 LO, 64 bits, Nikon Instruments), and was analyzed afterwards using Imaris (Bitplane). All imaging was performed at the Microscopy and Dynamic Imaging Unit of CNIC.
Quantification of primary granules
Blood samples were extracted every 4 h during 24 h from wild-type mice, starting at ZT1. Blood (100 μl) was RBC-lysed in hypotonic lysis buffer (0.15 M KH4Cl, 0.01 M KHCO3 and 0.01 M EDTA in water), washed with PBS and resuspended in 40 μl of PBS. Smears were then performed onto Superfrost Plus microscope slides (Thermo Scientific), air-flow dried and immediately fixed in 4% PFA for 10 min at room temperature. For the experiments with mutant mice, neutrophils were sorted as described, cytospun (600 r.p.m., medium acceleration) into microscope slides and fixed with 4% PFA for 10 min at room temperature. For CXCL2 stimulation experiments, sorted neutrophils were plated with RPMI medium on poly-l-lysine-covered 8-well μ-Slides (Ibidi) and left for 30 min to adhere. Cells were then incubated for 1 h with 50 ng ml−1 of recombinant CXCL2 (R&D Systems) or vehicle, and fixed with 4% PFA in PBS.
For neutrophils in blood versus tissues, we sorted cells and cytospun them as described. Before sorting, the tissues were extracted and kept in cold PBS (except liver, which was kept at room temperature in HBSS) and processed immediately after. To avoid activation of neutrophils, we did not perform digestion of the tissues, but mechanically dissociated them into single-cell suspensions by straining in 100-μm pore cell strainers (Falcon). Enrichment of leukocytes from liver was performed by 36% Percoll (GE Healthcare) in HBSS (Invitrogen) gradient centrifugation. Single-cell suspensions were incubated with antibodies against CD45, CD11b and Ly6G before sorting of CD45+, CD11b+ and Ly6G+ neutrophils. Finally, cytospun neutrophils were fixed as described above.
For the light cycle inversion experiments, WT mice were housed in light cabinets with inverted light cycle for 3 weeks before blood was withdrawn for analysis. Then, whole blood extracted in EDTA tubes was RBC-lysed and smears were performed as described above.
In all cases, after washing, cells were permeabilized for 30 min with 0.1% Triton, 25% FBS in PBS and stained with biotinylated anti-MPO at 4 °C overnight. Afterwards, cells were washed and incubated with A488-conjugated streptavidin for 3 h at room temperature. Finally, cells were washed, stained with 4,6-diamidino-2-phenylindole (DAPI) and mounted in Mowiol. Imaging was performed using a Leica SP8 X confocal microscopy system coupled to a DMI6000 inverted microscope, with ×100 (HC PL Apo CS2 100×/1.4 oil) magnification objective. Granule contents were analyzed using the Imaris software.
Ex vivo NET-formation assays
Neutrophils were sorted as previously described and 5 × 104 neutrophils were plated with RPMI medium on poly-l-lysine-covered 8-well μ-Slides (Ibidi), and left 30 min to adhere. Subsequently, cells were incubated for 2 h with 100 nM PMA or vehicle. Cells were then fixed using 4% PFA for 10 min, permeabilized with PBS with 0.1% Triton X-100, 1% goat serum plus 5% BSA and stained with antibodies against cit-H3, DNA (Sytox-green, Molecular Probes) and MPO. Whole-slide Z stack tilescan images were acquired with a Nikon A1R confocal system coupled to a Nikon Eclipse-Ti inverted microscope or a Leica SP5 confocal microscope, and analyzed using Imaris.
Antibody-induced ALI
A two-event model of transfusion-related ALI was adopted for our studies as described15. Male Balb/c mice (7–14-week-old) were injected intraperitoneally with 0.1 mg kg−1 LPS. At 24 h later, mice received an intravenous injection of 0.5 mg kg−1 anti-H2d (clone 34-1-2s; BioXCell). Some mice were treated 1 h before ALI induction with 12 mg kg−1 of chloramidine (Cayman Chemical) to block NET formation48. For survival experiments, mice were observed for 2 h during the acute phase of ALI.
Neutrophil elastase activity assay
Neutrophil elastase activity in plasma was measured using a commercially available kit (Neutrophil Elastase Activity Assay kit (Fluorometric), NAK246-1KT, Sigma-Aldrich) according to the manufacturer’s instructions. We analyzed 50 μl of plasma, with an excitation wavelength of 390 nm and emission wavelength of 510 nm in a Fluoroskan Ascent plate reader (Thermo Labsystems). Standard curve goodness-of-fit had an R2 of 0.99 in linear regression.
Intravital imaging of the lung
Intravital microscopy of the lung was performed as reported28. Briefly, mice were anesthetized and mechanically ventilated through the trachea using a small-animal ventilator model 687 (Harvard Apparatus). Then, right lateral thoracotomy was applied and the lung was positioned under the window of a custom-built fixation device. A mild vacuum was applied to hold the lung in position during microscopy. Mice were injected with AF647-conjugated Ly6G antibody to stain neutrophils, PE-conjugated CD41 antibody to stain platelets and Sytox-green (Molecular Probes) to stain extracellular DNA. Four-dimensional imaging was performed for each mouse in five random fields inside the visualization window, using the VIVO system12. Analysis was performed in Imaris software. For NET-like structures, we quantified the number of spots in the neutrophil extracellular DNA colocalization channel over time in all random fields.
Analysis of NET types in lungs
We found two types of NET-formation events in lungs during ALI. Flowing NETs were defined as events in which DNA was rapidly extruded out of the neutrophil and, most commonly, was washed away by the flow. Adherent NETs, which were defined as DNA being deployed slowly around the uropod region of the neutrophil as it crawled, stuck to the vasculature and were more resistant to blood-flow washing. To quantify the relative frequency of each of these events, we visually inspected every NET-formation event in our time-lapse footage and ascribed each event to one of the two categories, or to none if it was unclear.
Whole-mount immunostaining and tissue clearing
To confirm the presence and abundance of NETs in the lungs of mice after ALI induction we performed whole-mount immunostaining and tissue clearing of excised lungs. Mice were subject to ALI and euthanized with CO2 15 min after ALI induction. Mice were then perfused with 20 ml of saline through the left ventricle of the heart, and the lungs were collected in cold PBS. Afterwards, lungs were fixed at 4 °C overnight in PBS with 4% PFA and 30% sucrose. After three washes of 1 h with PBS at room temperature, tissues were permeabilized in methanol gradients in PBS for 30 min (PBS > 50% MeOH > 80% MeOH > 100% MeOH). Then, tissues were bleached with Dent’s bleach (15% H2O2, 16.7% DMSO in MeOH) for 1 h at room temperature, and then were rehydrated through descending methanol gradients in PBS (80% MeOH > 50% MeOH > PBS). Then tissues were incubated with blocking buffer containing PBS with 0.3% Triton X-100, 0.2% BSA, 5% DMSO, 0.1% azide and 25% FBS overnight at 4 °C with shaking. Afterwards, lungs were stained with antibodies against cit-H3, MPO and CD31 for 2 d at 4 °C with shaking. After washing for 24 h in washing buffer (PBS with 0.2% Triton X-100 and 3% NaCl), the tissues were stained with secondary antibodies anti-rabbit-AF647, anti-hamster-AF405 and streptavidin-AF488 for 24 h at 4 °C with shaking. After 24 h, tissues were washed for 24 h in washing buffer and were dehydrated in MeOH gradients in dH20 using glass containers for 30 min in each step (50% MeOH > 70% MeOH > 90% MeOH > 3× 100% MeOH). Finally, tissues were cleared for 30 min in MeOH with 50% BABB (benzyl alcohol:benzyl benzoate, 1:2) and afterwards in 100% BABB and imaged in a Leica SP8 X confocal microscopy system coupled to a DMI6000 inverted microscope.
CT for in vivo edema quantification
In vivo CT imaging was performed on a nanoPET/CT small-animal imaging system (Bioscan) equipped with a micro-focus X-ray source and a high-resolution detector (1,024 × 3,596 pixels; 48 μm pixel size). Acquisition parameters were 55 kVp and 73 µAs. Mice were intraperitoneally anesthetized and positioned onto a thermoregulated (30 °C) mouse bed until the scan was completed. An ophthalmic gel was administered in their eyes to prevent retinal drying. Six studies were acquired per animal: one prior the retro-orbital injection of anti-H2d to induce ALI, and five postinjection over 30 min approximately. Data were acquired in helical mode obtaining 240 projections per rotation with 768 μm pixel size. Reconstruction was performed with an FDK-based method included in the proprietary Nucline software (Medisoy). Pulmonary edema was quantified using Horos software (The Horos Project, distributed under LGPL license at Horosproject.org) as the change of the mean value, in Hounsfield units, within the region of interest of the lungs obtained by thresholding (−1,000 and −400 lower and upper thresholds, respectively) followed by a closing morphological operation.
Intravital imaging of the cremaster muscle to quantify platelet–neutrophil interactions
Intravital microscopy of the cremaster muscle after TNF-α stimulation (R&D Systems, 0.5 μg intrascrotal injection) was performed as previously reported12,15 using the VIVO system. A total of 10–20 venule segments per mouse were analyzed 150–210 min after TNF-α treatment in multiple fluorescence channels (Cy3/561 for PE, FITC/488 for fluorescein isothiocyanate (FITC) and Cy5/640 for allophycocyanin (APC)) and bright-field images with 1 × 1 or 2 × 2 binning with a 3-s interval for 2 min on each field of view. For double staining with phycoerythrin (PE)- and FITC-conjugated antibodies, acquisition was facilitated in single (FITC) and quadrant (PE) filters to avoid bleed-through of fluorescent signals between channels. For the visualization of leukocytes, fluorescently labeled anti-Ly6G-APC and anti-CD62L-FITC were injected, and to visualize platelets, anti-CD41-PE was injected intravenously at 1 μg per mouse. Quantification of neutrophil–platelet interactions in either the leading edge or trailing edge was performed using Slidebook (3i).
Circadian analysis of neutrophil phenotypes
Circadian blood samples were extracted every 4 h during 24 h from wild-type or experimental mice, starting at ZT1 (zeitgeber time, 1 h after the onset of light, 7:00 at the CNIC animal facility). For circadian surface marker analysis, blood counts were analyzed in an automated hemocytometer (Abacus Junior, Diatron), RBCs were lysed in hypotonic lysis buffer (0.15 M NH4Cl, 0.01 M KHCO3 and 0.01 M EDTA in water) and incubated 15 min with 0.25 μg of anti-Ly6G, anti-CD62L and anti-CXCR2, washed and analyzed in a Sony SP6800 Spectral Analyzer. Analysis was performed using FlowJo v.X (Tree Star).
Vascular permeability assay
A 0.5% solution of Evans blue in sterile PBS was prepared and 200 μl of the solution was injected intravenously into mice 15 min before transfusion-related ALI induction (or just LPS injection as control, see Antibody-induced ALI for the protocol). Mice were sacrificed 15 min later, and tissues extracted and weighed. Then, tissues were submerged in 0.5 ml formamide and incubated at 55 °C for 24 h. Tissues were removed and the tubes centrifuged for 5 min at 645g. Finally, supernatants were measured for absorbance at 610 nm using an xMark Microplate Spectrophotometer (BioRad) plate reader.
Human studies
We performed human proteomics, RNA-sequencing, NET-formation assays and granule quantification (see individual methods sections) in neutrophils from blood samples of heathy volunteers obtained at 8:00, 14:00 and 19:00 at Hospital 12 de Octubre. The study complied with all ethical regulations and was approved by the Ethical Research Committee of Hospital 12 de Octubre in Madrid (CEIm no. 18/389), and informed consent was obtained from all volunteers.
Human neutrophil granule quantification
Diurnal blood samples were extracted from healthy volunteers at 8:00, 14:00 and 19:00. Blood (100 μl) was processed as indicated for mouse neutrophils.
Human neutrophil isolation
Freshly obtained blood (12 ml) extracted from healthy volunteers at the indicated times were used to isolate human neutrophils using Histopaque-1119 and -1077 (Sigma) gradients. In a 50-ml Falcon tube, 15 ml of Histopaque-1119 was added and then 15 ml of Histopaque-1077 was carefully layered on top. Whole blood was carefully layered on top of the low-density solution, and tubes were centrifuged for 30 min at 340g at room temperature, with no brakes or acceleration. Neutrophils layering between the 1077 and 1119 interphase were carefully aspirated and the isolated neutrophils washed twice in PBS. We obtained over 95% purity as assessed by flow cytometry.
RNA-sequencing of human neutrophils
Total RNA was prepared with the RNA Extraction RNeasy Plus Mini-kit (Qiagen). Total RNA (1 ng) was used to generate barcoded RNA-seq libraries as previously described12.
Libraries were sequenced on a HiSeq 2500 (Illumina) to generate 60-base single reads. FastQ files for each sample were obtained using bcl2fastq v.2.20 software (Illumina). The RNA-sequencing experiments were performed at the Genomics Unit at CNIC.
Human ex vivo NET-formation assay
Human neutrophils (4 × 104) isolated as previously described were plated with RPMI medium on poly-l-lysine-covered 8-well μ-Slides (Ibidi) and left for 30 min to adhere. Adherent cells were incubated for 2 h with 250 ng ml−1 PMA or vehicle and then fixed using 4% PFA for 10 min, permeabilized with PBS and 0.1% Triton X-100, 1% goat serum plus 5% BSA and stained with antibodies against cit-H3 (Abcam), DNA (Sytox-green, Molecular Probes) and MPO (R&D Systems). Whole-slide Z stack tilescan images were acquired with a Leica SP5 confocal microscope. Analysis of extranuclear DNA was performed using ImageJ with an automated method for extranuclear DNA quantification made available at https://doi.org/10.6084/m9.figshare.7358642.v1.
Retrospective analysis of human ARDS
Data from ARDS were extracted from a prospective observational cohort study of 5,334 consecutive adult patients with community-acquired pneumonia admitted to the intensive care unit who developed ARDS between 1996 and 2016 (ref. 32), in the Hospital Clinic of Barcelona. The time of admission, severity and survival were determined for the 125 patients who developed ARDS. Patients were excluded if they had severe immunosuppression or active tuberculosis. ARDS was identified on the basis of the Berlin definition. Only time points with three or more data points were included in subsequent analyses. The study was approved by the ethics committee of Hospital Clinic of Barcelona (no. 2009/5451).
TEM imaging of neutrophils
Neutrophils from mouse and human blood were isolated as previously described (FACS sorting for mouse neutrophils, gradient isolation for human neutrophils), fixed in 4% PFA for 10 min and washed with PBS. Samples were then postfixed in distilled water containing 4% osmium tetroxide for 1 h at room temperature. After washing in distilled water, samples were stained in a block with 0.5% uranyl acetate in water for 10 min. Then, samples were dehydrated in ascending gradients of ethanol in water (30%, 50%, 70%, 95% and 100%) with a last step in acetone. Samples were then included in epoxy resin (Durcupan) in ascending steps of resin:acetone (1:3, 3:1) and then pure resin. Samples included in the resin were polymerized for 48 h at 60 °C and 60-nm slices were obtained in a Leica Ultracut S ultramicrotome in 200-mesh copper gratings. Finally, samples were counterstained with uranyl acetate and lead citrate. Images were acquired in a 100-kV Jeol JEM-1010 TEM microscope with a Gatan SC200 camera coupled to the system. Quantification of granule-covered cytoplasmic area was performed in ImageJ using a custom-made macro that allows calculation of cell, nucleus and granule areas, and which is made available at https://doi.org/10.6084/m9.figshare.7619093.v1.
Statistical analysis
Unless otherwise indicated, data are represented as mean values ± s.e.m. Paired or unpaired two-tailed t-test was used when two groups were compared, and comparison of more than two datasets was done using one-way analysis of variance (ANOVA) with Turkey’s posttest. Where applicable, normality was estimated using the D’Agostino–Pearson or Shapiro–Wilk normality test. Log-rank analysis was used for Kaplan–Meier survival curves. Sample exclusion was not performed unless evident signs of disease were found in a mouse. For determination of diurnal patterns, we performed COSINOR fitting of circadian curves12. To determine whether a diurnal curve displayed an oscillating pattern we used the COSINOR-calculated amplitudes and compared them with a hypothetical zero-amplitude curve (that is, with no circadian behavior) assuming that both curves have identical standard deviations. We finally compared the amplitudes of the two curves using unpaired two-tailed t-test analyses. All statistical analyses were performed using Prism v.7 (GraphPad Software), except proteomics and sequencing analysis, which were performed using R (see individual methods). A P value below 0.05 was considered statistically significant; nonsignificant differences are indicated accordingly.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Proteomics data for mouse and human neutrophils are available in the Peptide Atlas with accession number PASS01364. Proteomics data for Bmal1∆N mice at ZT5 and ZT13 and WT vehicle versus AMD3100-treated mice are also available in the Peptide Atlas with the accession number PASS01438. Mouse circadian transcriptomics used for the correlation analysis are available at the Gene Expression Omnibus with accession number GSE102310. Human sequencing data used for the correlation analysis are available at GEO with accession number GSE136632. All other data are available upon request.
Code availability
ImageJ macros for TEM granule quantification and extracellular DNA quantification are available in FigShare (see the relevant section of Methods for specific links). All other code is available upon request.
References
Cowland, J. B. & Borregaard, N. Granulopoiesis and granules of human neutrophils. Immunol. Rev. 273, 11–28 (2016).
Ley, K. et al. Neutrophils: new insights and open questions. Sci. Immunol. 3, eaat4579 (2018).
Borregaard, N., Sørensen, O. E. & Theilgaard-Mönch, K. Neutrophil granules: a library of innate immunity proteins. Trends Immunol. 28, 340–345 (2007).
Rørvig, S., Østergaard, O., Heegaard, N. H. & Borregaard, N. Proteome profiling of human neutrophil granule subsets, secretory vesicles, and cell membrane: correlation with transcriptome profiling of neutrophil precursors. J. Leukoc. Biol. 94, 711–721 (2013).
Borregaard, N. Neutrophils, from marrow to microbes. Immunity 33, 657–670 (2010).
Le Cabec, V., Cowland, J. B., Calafat, J. & Borregaard, N. Targeting of proteins to granule subsets is determined by timing and not by sorting: the specific granule protein NGAL is localized to azurophil granules when expressed in HL-60 cells. Proc. Natl Acad. Sci. USA 93, 6454–6457 (1996).
Evrard, M. et al. Developmental analysis of bone marrow neutrophils reveals populations specialized in expansion, trafficking, and effector functions. Immunity 48, 364–379.e8 (2018).
Grassi, L. et al. Dynamics of transcription regulation in human bone marrow myeloid differentiation to mature blood neutrophils. Cell Rep. 24, 2784–2794 (2018).
Tak, T., Tesselaar, K., Pillay, J., Borghans, J. A. & Koenderman, L. What’s your age again? Determination of human neutrophil half-lives revisited. J. Leukoc. Biol. 94, 595–601 (2013).
Casanova-Acebes, M. et al. Neutrophils instruct homeostatic and pathological states in naive tissues. J. Exp. Med. 215, 2778–2795 (2018).
He, W. et al. Circadian expression of migratory factors establishes lineage-specific signatures that guide the homing of leukocyte subsets to tissues. Immunity 49, 1175–1190.e7 (2018).
Adrover, J. M. et al. A neutrophil timer coordinates immune defense and vascular protection. Immunity 51, 966–967 (2019).
Grommes, J. & Soehnlein, O. Contribution of neutrophils to acute lung injury. Mol. Med. 17, 293–307 (2011).
Looney, M. R., Su, X., Van Ziffle, J. A., Lowell, C. A. & Matthay, M. A. Neutrophils and their Fcγ receptors are essential in a mouse model of transfusion-related acute lung injury. J. Clin. Investig. 116, 1615–1623 (2006).
Sreeramkumar, V. et al. Neutrophils scan for activated platelets to initiate inflammation. Science 346, 1234–1238 (2014).
Matthay, M. A. & Zemans, R. L. The acute respiratory distress syndrome: pathogenesis and treatment. Ann. Rev. Pathol. 6, 147–163 (2011).
Casanova-Acebes, M. et al. Rhythmic modulation of the hematopoietic niche through neutrophil clearance. Cell 153, 1025–1035 (2013).
Devi, S. et al. Neutrophil mobilization via plerixafor-mediated CXCR4 inhibition arises from lung demargination and blockade of neutrophil homing to the bone marrow. J. Exp. Med. 210, 2321–2336 (2013).
Zhang, D. et al. Neutrophil ageing is regulated by the microbiome. Nature 525, 528–532 (2015).
Papayannopoulos, V., Metzler, K. D., Hakkim, A. & Zychlinsky, A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J. Cell Biol. 191, 677–691 (2010).
Urban, C. F. et al. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog. 5, e1000639 (2009).
Csepregi, J. Z. et al. Myeloid-specific deletion of Mcl-1 yields severely neutropenic mice that survive and breed in homozygous form. J. Immunol. 201, 3793–3803 (2018).
Looney, M. R. et al. Platelet depletion and aspirin treatment protect mice in a two-event model of transfusion-related acute lung injury. J. Clin. Investig. 119, 3450–3461 (2009).
Caudrillier, A. et al. Platelets induce neutrophil extracellular traps in transfusion-related acute lung injury. J. Clin. Investig. 122, 2661–2671 (2012).
Hidalgo, A. et al. Heterotypic interactions enabled by polarized neutrophil microdomains mediate thromboinflammatory injury. Nat. Med. 15, 384–391 (2009).
Thomas, G. M. et al. Extracellular DNA traps are associated with the pathogenesis of TRALI in humans and mice. Blood 119, 6335–6343 (2012).
Looney, M. R., Gilliss, B. M. & Matthay, M. A. Pathophysiology of transfusion-related acute lung injury. Curr. Opin. Hematol. 17, 418–423 (2010).
Looney, M. R. et al. Stabilized imaging of immune surveillance in the mouse lung. Nat. Methods 8, 91–96 (2011).
Muller, J. E. et al. Circadian variation in the frequency of onset of acute myocardial infarction. N. Engl. J. Med. 313, 1315–1322 (1985).
Scheiermann, C., Kunisaki, Y. & Frenette, P. S. Circadian control of the immune system. Nat. Rev. Immunol. 13, 190–198 (2013).
Suarez-Barrientos, A. et al. Circadian variations of infarct size in acute myocardial infarction. Heart 97, 970–976 (2011).
Cilloniz, C. et al. Acute respiratory distress syndrome in mechanically ventilated patients with community-acquired pneumonia. Eur. Respir. J. 51, 1702215 (2018).
Faurschou, M. & Borregaard, N. Neutrophil granules and secretory vesicles in inflammation. Microbes Infect. 5, 1317–1327 (2003).
Meyer-Hoffert, U. Neutrophil-derived serine proteases modulate innate immune responses. Front. Biosci. (Landmark Ed.) 14, 3409–3418 (2009).
Pham, C. T. Neutrophil serine proteases: specific regulators of inflammation. Nat. Rev. Immunol. 6, 541–550 (2006).
Houghton, A. M. et al. Neutrophil elastase-mediated degradation of IRS-1 accelerates lung tumor growth. Nat. Med. 16, 219–223 (2010).
Talukdar, S. et al. Neutrophils mediate insulin resistance in mice fed a high-fat diet through secreted elastase. Nat. Med. 18, 1407–1412 (2012).
Coffelt, S. B. et al. IL-17-producing γδ T cells and neutrophils conspire to promote breast cancer metastasis. Nature 522, 345–348 (2015).
Quail, D. F. et al. Obesity alters the lung myeloid cell landscape to enhance breast cancer metastasis through IL5 and GM-CSF. Nat. Cell. Biol. 19, 974–987 (2017).
Doerschuk, C. M. The role of CD18-mediated adhesion in neutrophil sequestration induced by infusion of activated plasma in rabbits. Am. J. Respir. Cell Mol. Biol. 7, 140–148 (1992).
Yipp, B. G. et al. The lung is a host defense niche for immediate neutrophil-mediated vascular protection. Sci. Immunol. 2, eaam8929 (2017).
Muller, J. E. & Tofler, G. H. Circadian variation and cardiovascular disease. N. Engl. J. Med. 325, 1038–1039 (1991).
Engelmann, B. & Massberg, S. Thrombosis as an intravascular effector of innate immunity. Nat. Rev. Immunol. 13, 34–45 (2013).
Jimenez-Alcazar, M. et al. Host DNases prevent vascular occlusion by neutrophil extracellular traps. Science 358, 1202–1206 (2017).
Navarro, P. et al. General statistical framework for quantitative proteomics by stable isotope labeling. J. Proteome Res. 13, 1234–1247 (2014).
Garcia-Marques, F. et al. A novel systems-biology algorithm for the analysis of coordinated protein responses using quantitative proteomics. Mol. Cell. Proteomics 15, 1740–1760 (2016).
Trevisan-Herraz, M. et al. SanXoT: a modular and versatile package for the quantitative analysis of high-throughput proteomics experiments. Bioinformatics 35, 1594–1596 (2019).
Li, P. et al. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J. Exp. Med. 207, 1853–1862 (2010).
Acknowledgements
We thank members of the Comparative Medicine Unit and Advanced Microscopy Unit at CNIC. This study was supported by Intramural grants from the Severo Ochoa program (IGP-SO), a grant from Fundació La Marató de TV3 (120/C/2015-20153032), grant SAF2015-65607-R from Ministerio de Ciencia, Investigacion y Universidades (MCIU) with cofunding from Fondo Europeo de Desarrollo Regional, grant RTI2018-095497-B-I00 from MCIU and HR17_00527 from Fundación La Caixa (to A.H.), and fellowship BES-2013-065550 from MCIU (to J.M.A.), fellowship from La Caixa Foundation (ID 100010434, code LCF/BQ/DR19/11740022, to A.A.-C.) and fellowship Health-PERIS 2016–2020 (to C.C.) Funds were also obtained from Instituto de Salud Carlos III (FIS PI17/01601, to I.L.) and SAF2015-68632-R from MCIU (to M.A.M.); Wellcome Trust Seed Award in Science (206103/Z/17/Z, to D.R.), SFB1123-A1/A10 from Deutsche Forschungsgemeinschaft and ERC-AdG 692511 (to C.W.); SAF2017-84494-C2-R and Programa Red Guipuzcoana de Ciencia, Tecnología e Información 2018-CIEN-000058-01 (to J.R.-C.). Work at CIC biomaGUNE was performed under the Maria de Maeztu Units of Excellence Program from the Spanish State Research Agency (MDM-2017-0720). C.W. is a van de Laar professor of atherosclerosis. The CNIC is supported by the MCIU and the Pro-CNIC Foundation, and is a Severo Ochoa Center of Excellence (MEIC award SEV-2015-0505).
Author information
Authors and Affiliations
Contributions
J.M.A., A.A.-C, G.C., E.B.-K., E.C. and Y.R.-V. performed experiments. J.M.A., D.R. and A.R.-P. performed bioinformatic analysis. F.O. obtained human samples. C.W., M.A.M., J.R.-C., I.L. and J.V. contributed essential reagents, equipment, expertise and funds. C.C. and A.T. contributed clinical data. J.M.A. and A.H. designed and supervised experiments, and wrote the manuscript, which was edited by all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Ioana Visan was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Validation and analysis of neutrophil proteomics.
a, Experimental strategy for proteomic analysis of day-like (from P- and E-selectin treated mice) and night-like (from AMD3100-treated mice) neutrophils isolated by negative selection (see methods) from blood. b, Intracellular staining of proteins from the proteomics dataset for validation in fresh (blue) and aged (violet) neutrophils obtained as indicated in a. All the proteins analyzed correlated with the proteomics data; n = 3 mice per condition. c, GO terms of the differentially expressed proteins (FDR<0.05, see methods section for 18O proteomics) in the proteomics dataset, showing terms with p < 0.05 (from single samples of 60 million neutrophils pooled from 9 mice (night) and 6 mice (day)). Bubble size represents overlap of query vs. the GO term. d, Scatterplot, correlation coefficient and significance level (pvalue) of the Spearman’s correlation of the direction of change of common proteins and genes from our proteomic analysis of fresh and aged neutrophils (this paper) and circadian RNA-sequencing data previously reported (Adrover et al. 2019, from 3 mice at ZT5 and 3 mice at ZT13), showing poor correlation of RNA and protein content. e, Venn-diagram showing the number of differentially detected proteins (p < 0.05, see methods section for mouse TMT proteomics) between vehicle- and AMD3100-treated mice (at ZT5); n = 3 samples per group. f, Heatmap showing levels of granule proteins in this dataset, Note increased detection of most granule proteins in neutrophils from AMD3100-treated mice. Data in (b) are shown as mean ± SEM. *; p < 0.05; **, p < 0.01; ***, p < 0.001, as determined by unpaired two-tailed t-test analysis.
Extended Data Fig. 2 Degranulation of neutrophils in the circulation and in tissues.
a, Reactome pathway analysis of the proteome of night and day neutrophils (see methods section for 18O proteomics, from single samples of 60 million neutrophils pooled from 9 mice (night) and 6 mice (day)) showing pathways with p-value < 0.05. b, Light-scattering properties (a measure of granularity) of blood neutrophils during a full diurnal cycle, measured as side-scatter in flow cytometry. Data are for WT, CXCR2-, CXCR4- or Bmal1-deficient neutrophils, showing that cell-intrinsic disruption of clock regulators blunts diurnal fluctuation in granularity. Curves are repeated for two cycles (dashed line) to better appreciate the circadian pattern; n = 10 (WT), 3 (CXCR2∆N), 4 (CXCR4∆N) and 4 (Bmal1∆N) mice per time point. c, Kinetics of total (top) or aged (bottom) neutrophils in blood indicating times of release of young or accumulation of aged neutrophils; n = 5 mice (ZT13, ZT17, ZT21, ZT1 and ZT9), n = 4 mice (ZT5). d, Shift of the light cycle alters the pattern of granule content in neutrophils. Left, representative confocal images of sorted neutrophils (MPO, green; DAPI, blue; scale, 5 μm); right, granule content per cell at the indicated times and light regime; n = 3 mice. LD, light-dark cycle; DL, dark-light (inverted) cycle. e, Representative confocal images (scale, 1 μm) and f, quantification of granule content in neutrophils from the blood or tissues of WT mice, showing reduced granule counts in tissues compared with blood; n = 30 (blood, lung and spleen), n = 27 (Liver) cells from 3 mice. Data are shown as mean ± SEM. *; p < 0.05; ***, p < 0.001; n.s., not significant, as determined by one-way ANOVA with Dunnet’s multiple comparison test (d), or using the amplitude vs. zero two-tailed t-test for circadian curves (b).
Extended Data Fig. 3 CXCR2-deficient neutrophils are responsive to activating stimuli.
a, Representative confocal images (scale, 2 μm) and b, quantification of granule content (top) and MPO intensity (bottom) in CXCR2-deficient neutrophils upon LPS or PMA stimulation. Granule loss indicates that CXCR2-deficient neutrophils are responsive to inflammatory stimuli; n = 15 cells per group. Data are shown as mean ± SEM. **; p < 0.01; ***, p < 0.001, as determined.
Extended Data Fig. 4 Regulation of circadian patterns by Bmal1.
a, Representative confocal images (left) and quantification of granule content (right) in Bmal1-deficient neutrophils at ZT13 (night) and ZT5 (day). n = 30–31 cells from 3 mice; scale 2 μm. b, Ex vivo NET formation by Bmal1-deficient neutrophils at ZT5 and ZT13. Note that Bmal1-deficient neutrophils fail to display circadian oscillations in both granule and NET formation. n = 3 mice per time point. c, Experimental design of circadian proteomic analysis of Bmal1-deficient neutrophils. d, Granule proteins (left) and NET-associated proteins (right) in the circadian Bmal1∆N neutrophil proteome (n = 3 mice at ZT5 and n = 2 at ZT13). Black dots show all granule or NET-associated proteins, respectively, none of which reached significance in differential expression between night and day (FDR < 0.05, see methods section for TMT proteomics of mouse neutrophils). e, Heatmap of granule proteins in the circadian proteome of wild-type (same as in Fig. 1) and Bmal1∆N neutrophils. Note that the diurnal pattern is lost in Bmal1-deficient neutrophils. Data in (a-b) are shown as mean ± SEM; n.s., not significant, as determined by unpaired two-tailed t-test.
Extended Data Fig. 5 Normal circadian oscillations in Balb/c mice.
a, Total (left) and CD62LO aged (right) neutrophil counts in the blood of Balb/c mice; n = 4–5 mice per time. b, Circadian oscillations in CD62L and CXCR2 expression in neutrophils from Balb/c mice, measured as median fluorescence intensity (MFI) by flow cytometry n = 5 mice (ZT13, ZT17, ZT21, ZT1 and ZT9), n = 4 mice (ZT5). c, Side scatter values plotted together with surface levels of CD62L in neutrophils, showing similar fluctuation patterns as reported for C57BL/6 neutrophils; n = 5 mice (ZT13, ZT17, ZT21, ZT1 and ZT9), n = 4 mice (ZT5). All curves are repeated for two cycles (dashed line) to better appreciate the circadian pattern. Data are shown as mean ± SEM. P values were determined by the amplitude vs. zero two-tailed t-test.
Extended Data Fig. 6 Neutrophils and platelets in the lung microvasculature during ALI.
Quantification of neutrophil a, and platelet b, numbers per field of view over time in wild-type mice subject to ALI at night (ZT13, blue line) or during daytime (ZT5, red line), in the intravital imaging experiments shown in Fig. 4c. Insets show area under the curve (AUC) values; n = 15 fields from 4 mice in each group. c, Neutrophil numbers in the lungs of naïve, LPS-only and wild-type mice in which ALI was induced at ZT5 (n = 8 mice) or ZT13 (n = 5 mice), or at ZT5 in the presence of Cl-amidine (n = 5 mice), as determined by flow cytometry. Neutrophils d, and platelets e, numbers in mutant mice (Bmal1∆N purple line; CXCR4∆N blue line) from the intravital imaging experiments shown in Fig. 5d; n = 15 fields from 4 mice in each genotype. Insets show area under the curve (AUC) values. f, Neutrophil numbers in the lungs of LPS-only control mice (n = 5) or during ALI in Bmal1ΔN (n = 4 mice) or Cre- control (n = 4 mice); and CXCR4ΔN (n = 5 mice) or Cre- control mice (n = 4 mice), as determined by flow cytometry. g, Interactions between platelets and the uropod (U) or leading edge (LE) of adherent neutrophils, in the inflamed cremasteric microvessels of wild-type (n = 45 from 3 mice), Bmal1ΔN (n = 28 from 3 mice) and CXCR4ΔN mice (n = 31 from 3 mice); scale, 5 μm. Data are shown as mean ± SEM. **; p < 0.01; ***, p < 0.001; n.s., not significant, as determined by unpaired two-tailed t-test analysis (a-d) or one-way ANOVA with Dunnet’s multiple comparison test (e-g). In the insets in a-d, individual data points are not shown as this graph uses a mean ± SEM value for the area under the curve calculated from the data shown in the respective panels.
Extended Data Fig. 7 Vascular leakiness and types of NETs during ALI.
Vascular leakiness in a, wild-type, b, Bmal1∆N, and c, CXCR4∆N mice after induction of ALI (LPS + antibody) or in control mice treated with LPS only. WT and Bmal1∆N mice displayed increased leakiness only in lungs upon ALI induction, while CXCR4∆N mice were protected; n = 3 (LPS only) and 5 (ALI) mice per genotype; d, Time-lapse images showing examples of flowing and adherent NETs (asterisks) as observed by intravital imaging of the lung microvasculature during ALI, representative of n = 3 independent experiments. See also Supplementary Movie 5. e, Relative frequency of NET types in WT, Bmal1∆N and CXCR4∆N mice during ALI, n = 15 fields from 3 mice (WT), 10 fields from 3 mice (Bmal1∆N) and 15 fields from 3 mice (CXCR4∆N). Data are shown as mean ± SEM. *; p < 0.05; ***, p < 0.001; n.s., not significant, as determined by two-way ANOVA (a-c; unless otherwise specified, comparisons did not reach significance; and e).
Extended Data Fig. 8 Loss of circadian patterns in Bmal1∆N and CXCR4∆N mice.
a, Survival of wild-type, Bmal1∆N and CXCR4∆N mice subjected to ALI at night (ZT13, solid line) or daytime (ZT5, dashed line); n = 16 mice (ZT5) and 18 mice (ZT13) for wild-type, n = 10 mice per time point for Bmal1∆N; n = 12 mice (ZT5) and 14 mice (ZT13) for CXCR4∆N. b, Representative confocal images (top) and quantification of granule content (bottom) in CXCR4-deficient neutrophils at ZT13 and ZT5. Note the loss of diurnal fluctuations compared with wild-type mice (see Fig. 1e); n = 30 cells (from 3 mice) per time point; scale, 2 μm. c, Ex vivo NET formation after PMA stimulation by CXCR4∆N neutrophils analyzed at ZT13 (n = 3 mice) and ZT5 (n = 3 mice). Note the loss of diurnal changes in NET-formation compared with wild-type cells (see Fig. 2b); d, Neutrophil counts in blood at ZT5 and ZT13 in wild-type (n = 5 mice at ZT5 and 4 mice at ZT13) and Bmal1∆N mice (n = 4 mice per time point). Data are shown as mean ± SEM. *, p < 0.05; **, p < 0.01; n.s., not significant, as determined by two-sided log rank (Mantel-Cox) test (a) or unpaired two-tailed t-test (b-d).
Extended Data Fig. 9 Analysis of the human neutrophil proteome.
a, Experimental design. Blood from 10 healthy volunteers was extracted at 8 am, 2 pm and 7 pm. Neutrophils were purified for proteomic analysis, granule quantification and NET-formation assays. b, GO terms of the differentially expressed proteins between 8am and 2 pm in human neutrophils. c, Correlation analysis (Spearman) of the direction of change of common proteins and genes from paired human proteomic (n = 5 per time) and RNA sequencing (n = 3 per time) analysis, showing poor correlation of RNA and protein content. d, Volcano plot of the human neutrophil proteome highlighting proteins found in NETs. Red dots and labels show proteins that are significantly different among samples (p < 0.05), and black dots show all other NET proteins, and dots show the whole proteome dataset.
Extended Data Fig. 10 Graphical abstract.
Neutrophils are released from the bone marrow into the bloodstream enriched in granule-held antimicrobial, cytotoxic and NET-forming proteins. As they spend time in the circulation, they undergo a homeostatic process of proteome ‘disarming’ that is regulated by the clock gene Bmal1 and signaling through CXCR2. This process causes a reduction in granule content and their ability to form NETs, ultimately reducing their toxicity towards host tissues. During acute lung injury, the presence of granule-poor neutrophils at specific times of day or in CXCR4 mutants protects the lungs and increases survival. Disabling homeostatic degranulation in Bmal1 mutants, in contrast, increases organ damage and death at all times of day.
Supplementary information
Supplementary Video 1
Whole-mount immunostaining of clarified lungs showing almost complete absence of NETs in naïve lungs before ALI induction. Lungs were stained for MPO (neutrophils), DNA, citrunillated-histone 3 and CD31. NETs were defined as triple-positive MPO-DNA-citH3 events. Representative of three independently cleared lungs.
Supplementary Video 2
Whole-mount immunostaining of clarified lungs during ALI, showing abundant NETs scattered throughout the lung. Lungs were stained for MPO (neutrophils), DNA, citrunillated-histone 3 and CD31. NETs were defined as triple-positive MPO-DNA-citH3 events. Representative of three independently cleared lungs.
Supplementary Video 3
Intravital imaging of lungs during ALI, showing the formation of NET-like events in the pulmonary microvasculature. When ALI is induced at night (ZT13), more NET-like events (arrows) are produced compared with induction at noon (ZT5). Mice were treated intravenously with fluorescent anti-Ly6G to label neutrophils, and Sytox-green for extracellular DNA. NET-like events are defined as those in which DNA is extruded from a neutrophil. Representative of four independent mice per time.
Supplementary Video 4
Three-dimensional reconstruction of CT scans of lungs from WT, Bmal1∆N and CXCR4∆N mice before (t = 0) and after acute lung injury induction (t = 21). The appearance of edema is shown by fluid accumulation (red) in the lungs, and bones are shown for reference. Representative of seven independent mice per genotype.
Supplementary Video 5
Intravital microscopy of the lung microcirculation showing the two identified types of NETs produced by neutrophils during ALI: flowing NETs in which DNA is extruded out of the neutrophil body and is washed away by the blood flow; and adherent NETs, in which DNA is slowly extruded as the neutrophil crawls on the vessel, and adheres to the endothelial surface. Examples of both are shown. Representative of nine independent experiments.
Supplementary Video 6
Video abstract. Neutrophils are released from the bone marrow into the bloodstream enriched in granule-held antimicrobial, cytotoxic and NET-forming proteins. As they spend time in the circulation, they undergo a homeostatic process of proteome ‘disarming’ that is regulated by the clock gene Bmal1 and signaling through CXCR2. This process causes a reduction in granule content and their ability to form NETs, ultimately reducing their toxicity towards host tissues. During acute lung injury, the presence of granule-poor neutrophils at specific times of day or in CXCR4 mutants protects the lungs and increases survival. Disabling homeostatic degranulation in Bmal1 mutants, in contrast, increases organ damage and death at all times of day.
Supplementary Table
Supplementary Tables 1–8.
Rights and permissions
About this article
Cite this article
Adrover, J.M., Aroca-Crevillén, A., Crainiciuc, G. et al. Programmed ‘disarming’ of the neutrophil proteome reduces the magnitude of inflammation. Nat Immunol 21, 135–144 (2020). https://doi.org/10.1038/s41590-019-0571-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-019-0571-2
This article is cited by
-
Investigation of the rhythmic recruitment of tear neutrophils to the ocular surface and their phenotypes
Scientific Reports (2024)
-
Quantitative proteomics reveals tissue-specific, infection-induced and species-specific neutrophil protein signatures
Scientific Reports (2024)
-
The evolution and heterogeneity of neutrophils in cancers: origins, subsets, functions, orchestrations and clinical applications
Molecular Cancer (2023)
-
Human and mouse neutrophils share core transcriptional programs in both homeostatic and inflamed contexts
Nature Communications (2023)
-
Strategies of neutrophil diversification
Nature Immunology (2023)