Abstract
The cytokine interleukin (IL)-1β is a key mediator of antimicrobial immunity as well as autoimmune inflammation. Production of IL-1β requires transcription by innate immune receptor signaling and maturational cleavage by inflammasomes. Whether this mechanism applies to IL-1β production seen in T cell-driven autoimmune diseases remains unclear. Here, we describe an inflammasome-independent pathway of IL-1β production that was triggered upon cognate interactions between effector CD4+ T cells and mononuclear phagocytes (MPs). The cytokine TNF produced by activated CD4+ T cells engaged its receptor TNFR on MPs, leading to pro-IL-1β synthesis. Membrane-bound FasL, expressed by CD4+ T cells, activated death receptor Fas signaling in MPs, resulting in caspase-8-dependent pro-IL-1β cleavage. The T cell-instructed IL-1β resulted in systemic inflammation, whereas absence of TNFR or Fas signaling protected mice from CD4+ T cell-driven autoimmunity. The TNFR–Fas–caspase-8-dependent pathway provides a mechanistic explanation for IL-1β production and its consequences in CD4+ T cell-driven autoimmune pathology.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. Source data for Figs. 1–4 are provided with this paper.
References
Zheng, H. et al. Resistance to fever induction and impaired acute-phase response in interleukin-1 β-deficient mice. Immunity 3, 9–19 (1995).
Miller, L. S. et al. MyD88 mediates neutrophil recruitment initiated by IL-1R but not TLR2 activation in immunity against Staphylococcus aureus. Immunity 24, 79–91 (2006).
Jain, A., Song, R., Wakeland, E. K. & Pasare, C. T cell-intrinsic IL-1R signaling licenses effector cytokine production by memory CD4 T cells. Nat. Commun. 9, 3185 (2018).
Dinarello, C. A. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood 117, 3720–3732 (2011).
Broderick, L., De Nardo, D., Franklin, B. S., Hoffman, H. M. & Latz, E. The inflammasomes and autoinflammatory syndromes. Annu. Rev. Pathol. 10, 395–424 (2015).
Goodnow, C. C., Sprent, J., Fazekas de St Groth, B. & Vinuesa, C. G. Cellular and genetic mechanisms of self tolerance and autoimmunity. Nature 435, 590–597 (2005).
Masters, S. L., Simon, A., Aksentijevich, I. & Kastner, D. L. Horror autoinflammaticus: the molecular pathophysiology of autoinflammatory disease. Annu. Rev. Immunol. 27, 621–668 (2009).
Lopalco, G. et al. Interleukin-1 as a common denominator from autoinflammatory to autoimmune disorders: premises, perils, and perspectives. Mediators Inflamm. 2015, 194864 (2015).
Raphael, I., Nalawade, S., Eagar, T. N. & Forsthuber, T. G. T cell subsets and their signature cytokines in autoimmune and inflammatory diseases. Cytokine 74, 5–17 (2015).
Bluestone, J. A., Bour-Jordan, H., Cheng, M. & Anderson, M. T cells in the control of organ-specific autoimmunity. J. Clin. Invest. 125, 2250–2260 (2015).
Schenten, D. et al. Signaling through the adaptor molecule MyD88 in CD4+ T cells is required to overcome suppression by regulatory T cells. Immunity 40, 78–90 (2014).
Shaw, P. J., McDermott, M. F. & Kanneganti, T. D. Inflammasomes and autoimmunity. Trends Mol. Med. 17, 57–64 (2011).
Ippagunta, S. K. et al. Inflammasome-independent role of apoptosis-associated speck-like protein containing a CARD (ASC) in T cell priming is critical for collagen-induced arthritis. J. Biol. Chem. 285, 12454–12462 (2010).
Schott, W. H. et al. Caspase-1 is not required for type 1 diabetes in the NOD mouse. Diabetes 53, 99–104 (2004).
Cogswell, J. P. et al. NF-κB regulates IL-1β transcription through a consensus NF-κB binding site and a nonconsensus CRE-like site. J. Immunol. 153, 712–723 (1994).
Martinon, F., Burns, K. & Tschopp, J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell 10, 417–426 (2002).
Martin, B. N. et al. T cell-intrinsic ASC critically promotes TH17-mediated experimental autoimmune encephalomyelitis. Nat. Immunol. 17, 583–592 (2016).
Baldassare, J. J., Bi, Y. & Bellone, C. J. The role of p38 mitogen-activated protein kinase in IL-1β transcription. J. Immunol. 162, 5367–5373 (1999).
Geppert, T. D., Whitehurst, C. E., Thompson, P. & Beutler, B. Lipopolysaccharide signals activation of tumor necrosis factor biosynthesis through the Ras/Raf-1/MEK/MAPK pathway. Mol. Med. 1, 93–103 (1994).
Li, C. R., Mueller, E. E. & Bradley, L. M. Islet antigen-specific TH17 cells can induce TNF-⍺-dependent autoimmune diabetes. J. Immunol. 192, 1425–1432 (2014).
Franchi, L., Eigenbrod, T., Munoz-Planillo, R. & Nunez, G. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat. Immunol. 10, 241–247 (2009).
He, W. T. et al. Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res. 25, 1285–1298 (2015).
Bossaller, L. et al. Cutting edge: FAS (CD95) mediates noncanonical IL-1β and IL-18 maturation via caspase-8 in an RIP3-independent manner. J. Immunol. 189, 5508–5512 (2012).
Wang, L., Du, F. & Wang, X. TNF-⍺ induces two distinct caspase-8 activation pathways. Cell 133, 693–703 (2008).
Lemmers, B. et al. Essential role for caspase-8 in Toll-like receptors and NF-κB signaling. J. Biol. Chem. 282, 7416–7423 (2007).
Philip, N. H. et al. Activity of uncleaved caspase-8 controls anti-bacterial immune defense and TLR-induced cytokine production independent of cell death. PLoS Pathog. 12, e1005910 (2016).
Helft, J. et al. GM-CSF mouse bone marrow cultures comprise a heterogeneous population of CD11c+ MHCII+macrophages and dendriticcells. Immunity 42, 1197–1211 (2015).
Erlich, Z. et al. Macrophages, rather than DCs, are responsible for inflammasome activity in the GM-CSF BMDC model. Nat. Immunol. 20, 397–406 (2019).
Satpathy, A. T. et al. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J. Exp. Med. 209, 1135–1152 (2012).
Bachmann, M. F. & Kopf, M. On the role of the innate immunity in autoimmune disease. J. Exp. Med. 193, F47–F50 (2001).
Janeway, C. A.Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 54, 1–13 (1989).
VanderBorght, A., Geusens, P., Raus, J. & Stinissen, P. The autoimmune pathogenesis of rheumatoid arthritis: role of autoreactive T cells and new immunotherapies. Semin. Arthritis Rheum. 31, 160–175 (2001).
Fletcher, J. M., Lalor, S. J., Sweeney, C. M., Tubridy, N. & Mills, K. H. T cells in multiple sclerosis and experimental autoimmune encephalomyelitis. Clin. Exp. Immunol. 162, 1–11 (2010).
Pugliese, A. Autoreactive T cells in type 1 diabetes. J. Clin. Invest. 127, 2881–2891 (2017).
Mannie, M. D., Dinarello, C. A. & Paterson, P. Y. Interleukin 1 and myelin basic protein synergistically augment adoptive transfer activity of lymphocytes mediating experimental autoimmune encephalomyelitis in Lewis rats. J. Immunol. 138, 4229–4235 (1987).
Lin, C. C. & Edelson, B. T. New insights into the role of IL-1β in experimental autoimmune encephalomyelitis and multiple sclerosis. J. Immunol. 198, 4553–4560 (2017).
Adachi, M. et al. Enhanced and accelerated lymphoproliferation in Fas-null mice. Proc. Natl Acad. Sci. USA 93, 2131–2136 (1996).
Mande, P. et al. Fas ligand promotes an inducible TLR-dependent model of cutaneous lupus-like inflammation. J. Clin. Invest. 128, 2966–2978 (2018).
Orning, P. et al. Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death. Science 362, 1064–1069 (2018).
Conos, S. A., Lawlor, K. E., Vaux, D. L., Vince, J. E. & Lindqvist, L. M. Cell death is not essential for caspase-1-mediated interleukin-1β activation and secretion.Cell Death Differ. 23, 1827–1838 (2016).
Furlan, R. et al. Caspase-1 regulates the inflammatory process leading to autoimmune demyelination. J. Immunol. 163, 2403–2409 (1999).
De Jager, P. L. et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat. Genet. 41, 776–782 (2009).
Hinks, A. et al. Dense genotyping of immune-related disease regions identifies 14 new susceptibility loci for juvenile idiopathic arthritis. Nat. Genet. 45, 664–669 (2013).
International Multiple Sclerosis Genetics Consortium et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat. Genet. 45, 1353–1360 (2013).
Jin, Y. et al. Genome-wide association studies of autoimmune vitiligo identify 23 new risk loci and highlight key pathways and regulatory variants. Nat. Genet. 48, 1418–1424 (2016).
Tsoi, L. C. et al. Large scale meta-analysis characterizes genetic architecture for common psoriasis associated variants. Nat. Commun. 8, 15382 (2017).
Itoh, N. et al. Requirement of Fas for the development of autoimmune diabetes in nonobese diabetic mice. J. Exp. Med. 186, 613–618 (1997).
Kruglov, A. A., Lampropoulou, V., Fillatreau, S. & Nedospasov, S. A. Pathogenic and protective functions of TNF in neuroinflammation are defined by its expression in T lymphocytes and myeloid cells. J. Immunol. 187, 5660–5670 (2011).
Varanasi, V., Avanesyan, L., Schumann, D. M. & Chervonsky, A. V. Cytotoxic mechanisms employed by mouse T cells to destroy pancreatic β-cells. Diabetes 61, 2862–2870 (2012).
Acknowledgements
We thank all the members of the Pasare laboratory and R. Bagirzadeh for helpful discussions. We thank A. Ma for sharing WT and D117A Il1b constructs. We thank M. Jordan for sharing Zbtb46-GFP reporter mice. This work was supported by grants from the National Institutes of Health (AI113125 and AI123176) to C.P. M.M.M. was supported by a National Science Foundation Graduate Research Fellowship under grant 2017220107.
Author information
Authors and Affiliations
Contributions
C.P. and A.J. conceptualized the study, designed the experiments and wrote the manuscript. A.J. performed the majority of the experiments. R.A.I.C., M.M.M., A.S.C. and G.R.O. performed some of the experiments. J.D.K. and K.R.C. helped with EAE mouse experiments. A.V.C. provided Fasfl/fl mice. A.O. helped with experiments performed in Rip3−/− and Rip3−/−Casp8−/− mice. N.H.P. helped with Rip3−/−Casp8−/− DC experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Ioana Visan was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Representative Gating strategy for flow cytometric analysis.
a, Gating strategy for flow cytometric analysis of intracellular pro-IL-1β in CD11c+ DCs following BMDC-T cell co-culture. b, Gating strategy for flow cytometric analysis of CD11b+ monocytes and neutrophils frequency in various tissues.
Extended Data Fig. 2 Induction of pro-IL-1β in BMDCs is independent of TLR activation but dependent on TNF.
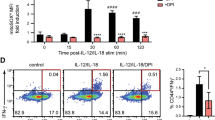
(a and b) Expression of intracellular pro-IL-1β measured by flow cytometry in WT, Tlr2−/−Tlr4−/−, or Myd88−/− BMDCs (live,CD90−CD11c+) cultured with TH0 cells in the presence of anti-CD3 Ab for 6 h. Fold change indicates proportion of pro-IL-1β+ BMDCs, compared to PBS controls. Error bars indicate s.e.m. three (n = 3) independent experiments. c, Expression of intracellular pro-IL-1β measured by flow cytometry in WT BMDCs (live, CD11c+) stimulated in vitro with recombinant TNF (20 ng/mL) for 6 h. Data are representative of two independent experiments. d, TNFα, as quantified by ELISA, in the supernatants of WT or Tnf−/− TH0 cells cultured with WT or Tnf−/− BMDCs in the presence of anti-CD3 Ab for 6 h. Error bars indicate s.e.m. from n = 4 independent experiments. e, lL-1β was quantified by ELISA in the supernatants of WT TH0 cells cultured with BMDCs of the indicated genotypes in the presence of anti-CD3 Ab and neutralizing CD40L Ab (10 µg/mL) for 6 h. Error bars indicate s.e.m. from n = 2 technical replicates. Data are representative of two independent experiments. (a, b, d) Statistical analysis was performed by paired, two-tailed Student’s t-test. *p < 0.05.
Extended Data Fig. 3 T cell-induced IL-1β production is only partially impaired in gasdermin-D deficient BMDCs.
a, lL-1β was quantified by ELISA in the supernatants of WT TH0 cells cultured with Il1b−/− BMDCs transduced with retrovirus expressing WT or D117A pro-IL-1β in the presence of anti-CD3 Ab for 18 h. Error bars indicate s.e.m. from n = 2 technical replicates. Data are representative of two independent experiments. b, lL-1β was quantified by ELISA in the supernatants of WT TH0 cells cultured with WT or Gsdmd−/− BMDCs in the presence of anti-CD3 Ab for 12 h. Error bars indicate s.e.m. from n = 6 independent experiments. Statistical analysis was performed by paired, two-tailed Student’s t-test. **p < 0.01.
Extended Data Fig. 4 T cells induce Fas-caspase-8 dependent death of interacting BMDCs.
a, Expression of surface FasL measured by flow cytometry in in vitro polarized TH1, TH2 and TH17 cells (live, CD90.2+) cultured with WT BMDCs in the presence of anti-CD3 Ab for 1 h. Data are representative of two independent experiments. (b and c) Cell death as assayed by Zombie Yellow viability dye was measured by flow cytometry in WT, Ripk3−/−, or Ripk3−/−Casp8−/− live,CD90.2+CD11c+ BMDCs cultured with WT TH0 cells in the presence or absence of anti-CD3 Ab and neutralizing TNF Ab (20μg/mL) or FasL Ab (10μg/mL) for 6 h. Data are representative of two independent experiments.
Extended Data Fig. 5 Gating strategy and post sort purity of various myeloid cell populations using in Fig. 5.
(a and b) Pre-sort and post-sort purity of FACS sorted BMDC subsets: CD11b+, CD11b+CD11c+MHCIIhi, or CD11b+CD11c+MHCIIint BMDCs b, and CD11c+MHCIIintCSF1R+ (GM-Macs) or CD11c+MHCIIhiCSF1R− (GM-DCs). Data are representative of 4 independent experiments. c, Post-sort purity of CD11c+ splenic DCs showing lack of CSF1R+ cell contamination. Data are representative of 4 independent experiments. d, Pre-sort and post-sort purity of CD11c+Zbtb46-GFP+Ly6C− splenic cDCs that were FACS sorted from pre-sorted total CD11c+ Zbtb46-GFP splenocytes to obtain a pure cDC population. Data are representative of three independent experiments.
Extended Data Fig. 6 Anti-CD3 stimulation of T cells in vivo leads to IL-1β dependent inflammatory cell recruitment dictated by Fas expression on CD11c+ cells.
a, Expression of intracellular pro-IL-1β measured by flow cytometry in CD11b+Ly6C−CSF1R+ macrophages, CD11b+Ly6G−Ly6Cint monocytes, CD11b+Ly6Chi inflammatory monocytes, and CD11b+Ly6G+ granulocytes quantified from the spleens of WT mice 3-4 h post anti-CD3 Ab injection (50μg, i.v.). n = 3 independent experiments are quantified in the inset. Statistical analysis was performed by paired, one-tailed Student’s t-test. b, Neutrophil infiltration as measured by flow cytometry in the spleen (left panel) or SI-LP (right panel) of WT mice 18 h post anti-CD3 Ab injection (20μg, i.p.). Error bars indicate s.e.m. from n = 4 independent experiments. c, qPCR of Il1b mRNA in lysates of splenocytes collected from WT or Rag1-/- mice 4 h post anti-CD3 Ab injection (50μg, i.v.). Data are normalized to Hprt. Error bars indicate s.e.m. from n = 3 technical replicates. Data are representative of two independent experiments. d, Neutrophil infiltration in the spleen of Rag1-/- mice as measured by flow cytometry 3-4 h post anti-CD3 Abinjection (50μg, i.v.). Error bars indicate s.e.m. from n = 3 independent experiments. e, Expression of Foxp3 and f, ICOS measured by flow cytometry in splenic live, CD4+ T cells from WT mice, 18 h post anti-CD3 Ab injection (20μg, i.p.). Error bars indicate s.e.m. from n = 3 independent experiments. g, Infiltration of CD11b+ cells as measured by flow cytometry in the SI-LP or (h) spleen of WT and Il1b-/- mice, 18 h post anti-CD3 Ab injection (20μg, i.p.). Error bars indicate s.e.m. from n = 3 independent experiments. i, Expression of cell surface Fas measured by flow cytometry in CD11b+ or CD11c+ BMDCs from given genotypes. Data are representative of two independent experiments. j, Neutrophil infiltration (live, CD11b+F480−) as measured by flow cytometry in the SI-LP of Fasfl/fl or Fasfl/fl x CD11c-cre (FasΔDC) mice, 18 h post anti-CD3 Ab injection (20μg, i.p.). Error bars indicate s.e.m. from n = 5 independent experiments. Splenocytes were taken from WT mice immunized with MOG35-55, and stimulated for 4 d in vitro with MOG in the presence of IL-1β, IL-23, and anti-IFNγ. k, lL-1β was quantified by ELISA in the supernatants of WT BMDCs cultured with CD4+ T cells isolated from the stimulated splenocyte culture in the presence of MOG35-55 and neutralizing antibodies TNF Ab (20μg/mL) or FasL Ab (10μg/mL) for 24 h. Error bars indicate s.e.m. from n = 2 independent experiments. (b-h,j) Statistical analysis was performed by unpaired, one-tailed Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, n.s. = not significant.
Extended Data Fig. 7 Illustration of “T cell-instructed” IL-1β production by MPs and its comparison to inflammasome induced IL-1β production by macrophages.
(Left) During inflammasome activation in monocytes and macrophages, TLR and NLR−caspase-1 activation leads to synthesis and cleavage of pro-IL-1β, respectively. Robust production of IL-1β as a result of inflammasome activation is critical for pathogen clearance. (Right) In contrast, “T cell-instructed” IL-1β production by antigen presenting MPs utilizes TNFR signaling for pro-IL1β synthesis while the cleavage signal is provided by Fas-caspase-8 axis. The IL-1β produced upon such T cell instruction drives cytokine production by effector CD4 T cells, systemic leukocyte recruitment, and autoimmunity. Figure was created using Biorender.
Supplementary information
Source data
Source Data Fig. 1
Unprocessed western blots
Source Data Fig. 2
Unprocessed western blots
Source Data Fig. 3
Unprocessed western blots
Source Data Fig. 4
Unprocessed western blots
Rights and permissions
About this article
Cite this article
Jain, A., Irizarry-Caro, R.A., McDaniel, M.M. et al. T cells instruct myeloid cells to produce inflammasome-independent IL-1β and cause autoimmunity. Nat Immunol 21, 65–74 (2020). https://doi.org/10.1038/s41590-019-0559-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-019-0559-y
This article is cited by
-
Recent advances and evolving concepts in Still’s disease
Nature Reviews Rheumatology (2024)
-
Dopamine signaling modulates microglial NLRP3 inflammasome activation: implications for Parkinson’s disease
Journal of Neuroinflammation (2022)
-
The potential convergence of NLRP3 inflammasome, potassium, and dopamine mechanisms in Parkinson’s disease
npj Parkinson's Disease (2022)
-
Insights into inflammasome regulation: cellular, molecular, and pathogenic control of inflammasome activation
Immunologic Research (2022)
-
The immune microenvironment features and response to immunotherapy in EBV-associated lymphoepithelioma-like cholangiocarcinoma
Hepatology International (2022)