Abstract
Lymph node fibroblastic reticular cells (FRCs) respond to signals from activated T cells by releasing nitric oxide, which inhibits T cell proliferation and restricts the size of the expanding T cell pool. Whether interactions with FRCs also support the function or differentiation of activated CD8+ T cells is not known. Here we report that encounters with FRCs enhanced cytokine production and remodeled chromatin accessibility in newly activated CD8+ T cells via interleukin-6. These epigenetic changes facilitated metabolic reprogramming and amplified the activity of pro-survival pathways through differential transcription factor activity. Accordingly, FRC conditioning significantly enhanced the persistence of virus-specific CD8+ T cells in vivo and augmented their differentiation into tissue-resident memory T cells. Our study demonstrates that FRCs play a role beyond restricting T cell expansion—they can also shape the fate and function of CD8+ T cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All data that relate to or support the observations described in this study are available from the corresponding authors upon reasonable request. The ATAC-seq and RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) database under accession no. GSE136905. The Microarray data are accessible through GEO accession no. GSE136958.
References
Mueller, S. N. & Germain, R. N. Stromal cell contributions to the homeostasis and functionality of the immune system. Nat. Rev. Immunol. 9, 618–629 (2009).
Turley, S. J., Fletcher, A. L. & Elpek, K. G. The stromal and haematopoietic antigen-presenting cells that reside in secondary lymphoid organs. Nat. Rev. Immunol. 10, 813–825 (2010).
Roozendaal, R. & Mebius, R. E. Stromal cell–immune cell interactions. Annu. Rev. Immunol. 29, 23–43 (2011).
Malhotra, D. et al. Transcriptional profiling of stroma from inflamed and resting lymph nodes defines immunological hallmarks. Nat. Immunol. 13, 499–510 (2012).
Malhotra, D., Fletcher, A. L. & Turley, S. J. Stromal and hematopoietic cells in secondary lymphoid organs: partners in immunity. Immunol. Rev. 251, 160–176 (2013).
Luther, S. A., Vogt, T. K. & Siegert, S. Guiding blind T cells and dendritic cells: a closer look at fibroblastic reticular cells found within lymph node T zones. Immunol. Lett. 138, 9–11 (2011).
Perez-Shibayama, C., Gil-Cruz, C. & Ludewig, B. Fibroblastic reticular cells at the nexus of innate and adaptive immune responses. Immunol. Rev. 289, 31–41 (2019).
Alexandre, Y. O. & Mueller, S. N. Stromal cell networks coordinate immune response generation and maintenance. Immunol. Rev. 283, 77–85 (2018).
Brown, F. D. & Turley, S. J. Fibroblastic reticular cells: organization and regulation of the T lymphocyte life cycle. J. Immunol. 194, 1389–1394 (2015).
Fletcher, A. L., Acton, S. E. & Knoblich, K. Lymph node fibroblastic reticular cells in health and disease. Nat. Rev. Immunol. 15, 350–361 (2015).
Chang, J. E. & Turley, S. J. Stromal infrastructure of the lymph node and coordination of immunity. Trends Immunol. 36, 30–39 (2015).
Luther, S. A., Tang, H. L., Hyman, P. L., Farr, A. G. & Cyster, J. G. Coexpression of the chemokines ELC and SLC by T zone stromal cells and deletion of the ELC gene in the plt/plt mouse. Proc. Natl Acad. Sci. USA 97, 12694–12699 (2000).
Bajenoff, M. et al. Stromal cell networks regulate lymphocyte entry, migration, and territoriality in lymph nodes. Immunity 25, 989–1001 (2006).
Asperti-Boursin, F., Real, E., Bismuth, G., Trautmann, A. & Donnadieu, E. CCR7 ligands control basal T cell motility within lymph node slices in a phosphoinositide 3-kinase independent manner. J. Exp. Med. 204, 1167–1179 (2007).
Worbs, T., Mempel, T. R., Bolter, J., von Andrian, U. H. & Forster, R. CCR7 ligands stimulate the intranodal motility of T lymphocytes in vivo. J. Exp. Med. 204, 489–495 (2007).
Denton, A. E., Roberts, E. W., Linterman, M. A. & Fearon, D. T. Fibroblastic reticular cells of the lymph node are required for retention of resting but not activated CD8(+) T cells. Proc. Natl Acad. Sci. USA 111, 12139–12144 (2014).
Schumann, K. et al. Immobilized chemokine fields and soluble chemokine gradients cooperatively shape migration patterns of dendritic cells. Immunity 32, 703–713 (2010).
Marsland, B. J. et al. CCL19 and CCL21 induce a potent proinflammatory differentiation program in licensed dendritic cells. Immunity 22, 493–505 (2005).
Link, A. et al. Fibroblastic reticular cells in lymph nodes regulate the homeostasis of naive T cells. Nat. Immunol. 8, 1255–1265 (2007).
Acton, S. E. et al. Dendritic cells control fibroblastic reticular network tension and lymph node expansion. Nature 514, 498–502 (2014).
Astarita, J. L. et al. The CLEC-2-podoplanin axis controls the contractility of fibroblastic reticular cells and lymph node microarchitecture. Nat. Immunol. 16, 75–84 (2015).
Lukacs-Kornek, V. et al. Regulated release of nitric oxide by nonhematopoietic stroma controls expansion of the activated T cell pool in lymph nodes. Nat. Immunol. 12, 1096–1104 (2011).
Siegert, S. et al. Fibroblastic reticular cells from lymph nodes attenuate T cell expansion by producing nitric oxide. PloS ONE 6, e27618 (2011).
Khan, O. et al. Regulation of T cell priming by lymphoid stroma. PloS ONE 6, e26138 (2011).
Cremasco, V. et al. B cell homeostasis and follicle confines are governed by fibroblastic reticular cells. Nat. Immunol. 15, 973–981 (2014).
Zhang, Y. et al. Plasma cell output from germinal centers is regulated by signals from Tfh and stromal cells. J. Exp. Med. 215, 1227–1243 (2018).
Huang, H. Y. et al. Identification of a new subset of lymph node stromal cells involved in regulating plasma cell homeostasis. Proc. Natl Acad. Sci. USA 115, E6826–E6835 (2018).
Brown, G. C. Regulation of mitochondrial respiration by nitric oxide inhibition of cytochrome c oxidase. Biochim. Biophys. Acta Bioenerg. 1504, 46–57 (2001).
Wink, D. A. et al. DNA deaminating ability and genotoxicity of nitric-oxide and its progenitors. Science 254, 1001–1003 (1991).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Buck, M. D., O’Sullivan, D. & Pearce, E. L. T cell metabolism drives immunity. J. Exp. Med. 212, 1345–1360 (2015).
MacIver, N. J., Michalek, R. D. & Rathmell, J. C. Metabolic regulation of T lymphocytes. Annu. Rev. Immunol. 31, 259–283 (2013).
Chang, C.-H. et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251 (2013).
Wang, R. N. et al. The transcription factor myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882 (2011).
Tyrakis, P. A. et al. S-2-hydroxyglutarate regulates CD8(+) T-lymphocyte fate. Nature 540, 236–241 (2016).
Finlay, D. K. et al. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8(+) T cells. J. Exp. Med. 209, 2441–2453 (2012).
Roychoudhuri, R. et al. BACH2 regulates CD8(+) T cell differentiation by controlling access of AP-1 factors to enhancers. Nat. Immunol. 17, 851–860 (2016).
Li, Q. S., Eppolito, C., Odunsi, K. & Shrikant, P. A. Antigen-induced Erk1/2 activation regulates Ets-1-mediated sensitization of CD8+ T cells for IL-12 responses. J. Leukoc. Biol. 87, 257–263 (2010).
Kurachi, M. et al. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8(+) T cells. Nat. Immunol. 15, 373–383 (2014).
Xin, G. et al. A critical role of IL-21-induced BATF in sustaining CD8-T-cell-mediated chronic viral control. Cell Rep. 13, 1118–1124 (2015).
Gray, S. M., Amezquita, R. A., Guan, T. X., Kleinstein, S. H. & Kaech, S. M. Polycomb repressive complex 2-mediated chromatin repression guides effector CD8(+) T cell terminal differentiation and loss of multipotency. Immunity 46, 596–608 (2017).
Sen, D. R. et al. The epigenetic landscape of T cell exhaustion. Science 354, 1165–1169 (2016).
Portt, L., Norman, G., Clapp, C., Greenwood, M. & Greenwood, M. T. Anti-apoptosis and cell survival: a review. Biochim. Biophys. Acta Mol. Cell Res. 1813, 238–259 (2011).
Scott-Browne, J. P. et al. Dynamic changes in chromatin accessibility occur in CD8(+) T cells responding to viral infection. Immunity 45, 1327–1340 (2016).
Yu, B. F. et al. Epigenetic landscapes reveal transcription factors that regulate CD8(+) T cell differentiation. Nat. Immunol. 18, 573–582 (2017).
Scharer, C. D., Bally, A. P. R., Gandham, B. & Boss, J. M. Cutting edge: chromatin accessibility programs CD8 T cell memory. J. Immunol. 198, 2238–2243 (2017).
van der Windt, G. J. W. et al. Mitochondrial respiratory capacity is a critical regulator of CD8(+) T cell memory development. Immunity 36, 68–78 (2012).
Pan, Y. D. et al. Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature 543, 252–256 (2017).
Hunter, C. A. & Jones, S. A. IL-6 as a keystone cytokine in health and disease. Nat. Immunol. 16, 448–457 (2015).
Kopf, M. et al. Impaired immune and acute-phase responses in interleukin-6-deficient mice. Nature 368, 339–342 (1994).
Ladel, C. H. et al. Lethal tuberculosis in interleukin-6-deficient mutant mice. Infect. Immun. 65, 4843–4849 (1997).
Lauder, S. N. et al. Interleukin-6 limits influenza-induced inflammation and protects against fatal lung pathology. Eur. J. Immunol. 43, 2613–2625 (2013).
Pasztoi, M. et al. Mesenteric lymph node stromal cell-derived extracellular vesicles contribute to peripheral de novo induction of Foxp3+ regulatory T cells. Eur. J. Immunol. 47, 2142–2152 (2017).
Fletcher, A. et al. Reproducible isolation of lymph node stromal cells reveals site-dependent differences in fibroblastic reticular cells. Front. Immunol. 2, 35 (2011).
Yu, M. et al. Fibroblastic reticular cells of the lymphoid tissues modulate T cell activation threshold during homeostasis via hyperactive cyclooxygenase-2/prostaglandin E-2 axis. Sci. Rep. 7, 3350 (2017).
Knoblich, K. et al. The human lymph node microenvironment unilaterally regulates T-cell activation and differentiation. PloS Biol. 16, 24 (2018).
Rodda, L. B. et al. Single-cell RNA sequencing of lymph node stromal cells reveals niche-associated heterogeneity. Immunity 48, 1014–1028 (2018).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Liberzon, A. et al. The molecular signatures database hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Pircher, H., Burki, K., Lang, R., Hengartner, H. & Zinkernagel, R. M. Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature 342, 559–561 (1989).
Laidlaw, B. J. et al. Cooperativity between CD8+ T cells, non-neutralizing antibodies, and alveolar macrophages is important for heterosubtypic influenza virus immunity. PloS Pathog. 9, e1003207 (2013).
Mueller, S. N. et al. Qualitatively different memory CD8+ T cells are generated after lymphocytic choriomeningitis virus and influenza virus infections. J. Immunol. 185, 2182–2190 (2010).
Corces, M. R. et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 48, 1193–1203 (2016).
Dunham, I. et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Acknowledgements
This work was supported by the US National Institutes of Health (grant no. RO1 5R01DK074500 to S.J.T. and W.N.H.; no. P01AI108545 to A.H.S; no. T32 CA207021 to M.W.L) and by a Howard Hughes Medical Institute Gilliam Fellowship for Advanced Study (to F.D.B). We thank members of the Turley, Haining and Sharpe laboratories for scientific insights and discussions. We also thank A. E. Mayfield for critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
F.D.B. conceived and conducted most of the experiments, analyzed and interpreted data and wrote the manuscript. D.R.S. conducted experiments and analyzed and interpreted data. J.G., M.W.L., V.L.-K., F.A.S., H-J.K., K.B.Y., S.J.H.R., K.B. and V.N.K. conducted experiments and interpreted data. J.D.T. and I.A.S. discussed data and provided technical assistance. V.C., N.N.D. and B.D.M. discussed and interpreted data. A.H.S., W.N.H. and S.J.T. directed the study, analyzed and interpreted results and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
V.N.K. and S.J.T. are employees of Genentech. W.N.H. is an employee of Merck. V.C. is an employee of Novartis. F.D.B. is an employee of Neon Therapeutics. The authors declare no competing interests.
Additional information
Peer review information L. A. Dempsey was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
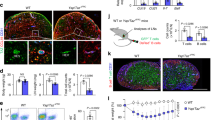
Supplementary Figure 1 FRCs dampen IFN-γ production yet enhance the production of IL-2 and TNF in newly activated CD8+ T cells.
(a) Gating strategy to highlight CD8+ T cells within whole splenocyte mixtures (24 h post anti-CD3/CD28 activation). (b) ICS of IFN-γ in CD8+ T cells activated in splenocyte mixtures via anti-CD3/CD28 for 24 h or activated via anti-CD3/CD28 for 24 h in the presence of wild-type FRCs. (c) Summary of results in b (n = 4 [Activated T cells] and n = 5 [Activated T cells + WT FRCs] biologically independent cell cultures per group). ****P < 0.0001 (two-tailed Student’s t-test). Data in c are composite of two biologically independent experiments (mean ± s.d). (d) ICS for IL-2 (top) and TNF (bottom) in purified CD8+ T cells activated via plate-bound anti-CD3 and soluble anti-CD28 for 24 h or activated via plate-bound anti-CD3 and soluble anti-CD28 in the presence of WT FRCs. (e) Summary of results in d as percentage of cytokine positive cells (n = 4 biologically independent cell cultures per group) **P = 0.0053 (IL-2), **P = 0.0018 (TNF) (two-tailed Student’s t-test) Data in e are a composite of two biologically independent experiments (mean ± s.d).
Supplementary Figure 2 CD8+ T cells activated alone or near Nos2−/− FRCs experience the same degree of initial activation and proliferation.
(a) CD25 expression on resting CD8+ T cells (resting), those activated in splenocyte mixtures via soluble anti-CD3/CD28 for 24 h (activated) or activated via anti-CD3/CD28 for 24 h in the presence of Nos2−/− FRCs. Top panel, representative plots; bottom panel, summary (n = 2 biologically independent cell cultures per group). (b) CFSE dilution in same CD8+ T cell conditions as a at 48 h post activation. Top panel, representative plots; bottom panel, summary (n = 3 biologically independent cell cultures per group). ns P = 0.2829 (two-tailed Student’s t-test). Data are representative of two biologically independent experiments (mean ± s.d).
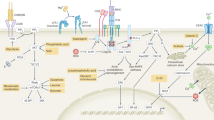
Supplementary Figure 3 Characteristics of chromatin accessible regions, RNA transcripts and epigenetic signatures in CD8+ T cells.
(a) Scatterplot displaying correlation in peak intensity between biological replicates for each CD8+ T cell condition. The conditions were CD8+ T cells sorted following activation in whole splenocyte mixture via soluble anti-CD3/CD28 for 48 h (Act), activated with anti-CD3/CD28 in presence of Nos2−/− FRCs (FRC) or activated with anti-CD3/CD28 plus 100ng/ml recombinant IL-6 (IL-6). (b) Principal component analysis of biological replicates for CD8+ T cells under each condition across all 81, 821 ChARs. (c) ATAC-seq fragment sizes for all three conditions (n = 3 biologically independent cell cultures per group). (d) Fold enrichment of regions annotated for histone marks and enhancer states for all three conditions. Data represent one experiment with three biologically independent cell cultures per group. (e) Scatterplot displaying correlation in gene expression between biological replicates for each CD8+ T cell condition. The conditions were CD8+ T cells sorted following activation in whole splenocyte mixtures via soluble anti-CD3/CD28 for 48 h (Act), activated with anti-CD3/CD28 in presence of Nos2−/− FRCs (FRC) or activated with anti-CD3/CD28 plus 100ng/ml recombinant IL-6 (IL-6). (f) Principal component analysis of biological replicates for CD8+ T cells under each condition. (n = 3 biologically independent cell cultures [FRC] and 2 biologically independent cell cultures [Act and IL-6]). (g) Display of gene expression using hierarchical clustering of replicates by condition with Pearson correlation. Data represent one experiment (n = 3 biologically independent cell cultures [FRC] and 2 biologically independent cell cultures [Act only and Act + IL-6]). (h) Overlap of H3K4me3 peaks identified in naive, terminal effector and memory precursor cells (Gray et al. 2017) with the FRC, IL-6 and Act conditions. (i) Overlap of ATAC-seq profiles corresponding to naive, effector and memory T cells (Sen et al. 2016) with the FRC, IL-6 and Act conditions.
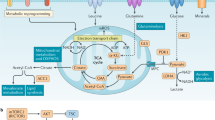
Supplementary Figure 4 IL-6 enhances phospho S6 levels and neutral lipid accumulation in activated T cells.
(a) Phospho flow cytometry of p-S6 levels in resting CD8+ T cells or those activated in whole splenocyte mixtures via anti-CD3/CD28 for 48 h or activated via anti-CD3/CD28 for 48 h in the presence of recombinant IL-6 (100ng/ml). Left panel, representative plot; right panel, summary (n = 6 biologically independent cell cultures per group) ****P < 0.0001 (two-tailed Student’s t-test) (b) Bodipy staining in CD8+ T cells either solely activated for 48 h with anti-CD3/CD28 or activated via anti-CD3/CD28 in the presence of recombinant IL-6 (100ng/ml). Left panel, representative plot; right panel, summary (n = 4 [Activated T cells] and 5 [Activated T cells + IL-6] biologically independent cell cultures) ns P = 0.0720 (two-tailed Student’s t-test) Data are a composite of two biologically independent experiments (mean ± s.d).
Supplementary Figure 5 FRCs extend the longevity of activated CD8+ T cells in vitro and during acute LCMV infection.
(a) Frequency of viable CD8+ T cells activated in splenocyte mixtures via soluble anti-CD3/CD28 for 48 h versus CD8+ T cells activated via anti-CD3/CD28 for 48 h in the presence of Nos2−/− FRCs. (b) Summary of results in a (n = 2 biologically independent cell cultures per group). (c) Frequency of viable T cells from a after being sorted and allowed to rest in alpha-mem media (supplemented with 10% fetal bovine serum and 1% penicillin streptomycin) for three days. (d) Summary of results in c (n = 2 biologically independent cell cultures per group). Data in b and d are representative of two biologically independent experiments. (e) Competitive frequencies of pre-activated P14 T cells before transfer (Day 0) and 7 days following transfer into wild type recipients infected with LCMV Armstrong. Transferred T cells were recovered from the lungs of infected animals. Top panel, representative plots; bottom panel, summary (n = 3 mice per group) *P = 0.0185 (two-tailed ratio paired t-test). (f) Frequency of competitive mixes at Day 0 and 7 days post LCMV infection. Transferred T cells were recovered from the spleen of infected animals. Top panel, representative plots; bottom panel, summary (n = 10 mice per group).****P < 0.0001 (two-tailed ratio paired t-test). Data in f are representative of three biologically independent experiments. (g) BrdU incorporation in transferred P14 T cells 8 days after the onset of influenza infection. Transferred T cells were recovered from the lungs of infected animals. (h) Summary of results in g (n = 5 mice per group). ns P = 0.4011 (two-tailed paired t-test) (mean ± s.d).
Supplementary Figure 6 Neutralizing IL-6 prior to adoptive transfer decreases the absolute number of recovered IL-2+ and TNF+ P14 T cells.
(a) Competitive frequencies of P14 T cells activated in whole splenocyte mixture with anti-CD3/CD28 in the presence of Nos2−/− FRCs and 10 μg/ml anti-IL-6 blocking antibody versus P14 T cells activated under the same conditions with 10 μg/ml of isotype control antibody. Il6−/− recipients were used for the transfers in a. Top panel, representative plots; bottom panel, summary. (n = 7 mice per group) ***P = 0.0004 (two-tailed ratio paired t-test) (b) Representative plots of ICS for IL-2 (left) and TNF (right) from CD8+ T cells recovered from the lungs of infected mice 9 days post influenza infection (n = 7 mice per group) (c) Summary of results in b (n = 7 mice per group) ns P = 0.0562 (IL-2), ns P = 0.6148 (TNF) (two-tailed paired t-test). (d) Absolute number of recovered cytokine positive P14 T cells. (n = 7 mice per group) *P = 0.0375 (IL-2), *P = 0.0265 (TNF) (two-tailed paired t-test), data in c shown as mean ± s.d. For b-d, the transferred cells were re-challenged in vitro with gp33 peptide (KAVYNFATC) for 6 h in the presence of Brefeldin A, followed by ICS staining to measure cytokine production.
Supplementary Figure 7 Generation and infection of bone marrow chimeras.
(a) Percent chimerism (following tail vein bleed) in C57BL/6J (WT) or Il6−/− host 6-12 weeks after transfer of donor cells. Representative plots shown in a. (b) Left panel, summary of data in a; right panel, hematopoietic cell lineage reconstitution in irradiated recipients. n = 10 mice per group (mean ± s.d) (c) Frequency of CD45− lymph node stromal cells. Top panel, representative plots; bottom panel, graphic display (n = pooled popliteal lymph nodes from 3 mice). (d) ICS for IL-6 in stromal populations from popliteal lymph nodes. The lymph nodes were harvested on day 8 following CFA/OVA immunization and OT-1 T cell injection. 100 μg Brefeldin A was injected intravenously (tail vein) to block protein transport 5 h prior to lymph node harvest and digest. Single cell suspensions were placed in Brefeldin A for an additional 2 h prior to ICS staining. Top panel, representative plots; bottom panel, graphic display. Data are representative of two biologically independent experiments with pooled popliteal lymph nodes from 3 wild-type C57BL/6J mice. (e) Frequency of CD44+, CD8+ T cells within the lungs of WT and Il6−/− bone marrow chimeras. Analysis conducted in animals that successfully cleared influenza infection from Fig. 7h (n = 6 of 10 mice for WT recipients and n = 1 of 10 mice for Il6−/− recipients) (mean ± SEM).
Supplementary information
Supplementary Information
Supplementary Figs. 1–7.
Rights and permissions
About this article
Cite this article
Brown, F.D., Sen, D.R., LaFleur, M.W. et al. Fibroblastic reticular cells enhance T cell metabolism and survival via epigenetic remodeling. Nat Immunol 20, 1668–1680 (2019). https://doi.org/10.1038/s41590-019-0515-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-019-0515-x
This article is cited by
-
PI16+ reticular cells in human palatine tonsils govern T cell activity in distinct subepithelial niches
Nature Immunology (2023)
-
Fibroblasts in metastatic lymph nodes confer cisplatin resistance to ESCC tumor cells via PI16
Oncogenesis (2023)
-
The advent of immune stimulating CAFs in cancer
Nature Reviews Cancer (2023)
-
Effector and stem-like memory cell fates are imprinted in distinct lymph node niches directed by CXCR3 ligands
Nature Immunology (2021)
-
The mesenchymal context in inflammation, immunity and cancer
Nature Immunology (2020)