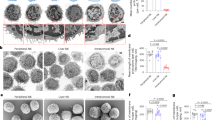
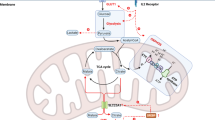
Abstract
Natural killer (NK) cells have crucial roles in tumor surveillance. We found that tumor-infiltrating NK cells in human liver cancers had small, fragmented mitochondria in their cytoplasm, whereas liver NK cells outside tumors, as well as peripheral NK cells, had normal large, tubular mitochondria. This fragmentation was correlated with reduced cytotoxicity and NK cell loss, resulting in tumor evasion of NK cell-mediated surveillance, which predicted poor survival in patients with liver cancer. The hypoxic tumor microenvironment drove the sustained activation of mechanistic target of rapamycin-GTPase dynamin-related protein 1 (mTOR-Drp1) in NK cells, resulting in excessive mitochondrial fission into fragments. Inhibition of mitochondrial fragmentation improved mitochondrial metabolism, survival and the antitumor capacity of NK cells. These data reveal a mechanism of immune escape that might be targetable and could invigorate NK cell-based cancer treatments.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Microarray data were deposited into the National Center for Biotechnology Information Gene Expression Omnibus repository (accession number: GSE120123). The clinical characteristics of all patients included in the present study are shown in Supplementary Tables 1–3. All gene sets are shown in Supplementary Table 4. The antibodies used are shown in Supplementary Table 5. Full scans of all of the blots and gels are included in the Source Data. The data that support the findings of this study are available from the corresponding author upon request.
References
Barry, K. C. et al. A natural killer–dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments. Nat. Med. 24, 1178–1191 (2018).
Lopez-Soto, A., Gonzalez, S., Smyth, M. J. & Galluzzi, L. Control of metastasis by NK cells. Cancer Cell 32, 135–154 (2017).
Morvan, M. G. & Lanier, L. L. NK cells and cancer: you can teach innate cells new tricks. Nat. Rev. Cancer 16, 7–19 (2016).
Dadi, S. et al. Cancer immunosurveillance by tissue-resident innate lymphoid cells and innate-like T cells. Cell 164, 365–377 (2016).
Ghesquiere, B., Wong, B. W., Kuchnio, A. & Carmeliet, P. Metabolism of stromal and immune cells in health and disease. Nature 511, 167–176 (2014).
Chang, C. H. et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell 162, 1229–1241 (2015).
Gemta, L. F. et al. Impaired enolase 1 glycolytic activity restrains effector functions of tumor-infiltrating CD8+ T cells. Sci. Immunol. 4, eaap9520 (2019).
Ma, C. et al. NAFLD causes selective CD4+ T lymphocyte loss and promotes hepatocarcinogenesis. Nature 531, 253–257 (2016).
Sugiura, A. & Rathmell, J. C. Metabolic barriers to T cell function in tumors. J. Immunol. 200, 400–407 (2018).
Assmann, N. et al. Srebp-controlled glucose metabolism is essential for NK cell functional responses. Nat. Immunol. 18, 1197–1206 (2017).
Marcais, A. et al. The metabolic checkpoint kinase mTOR is essential for IL-15 signaling during the development and activation of NK cells. Nat. Immunol. 15, 749–757 (2014).
Keating, S. E. et al. Metabolic reprogramming supports IFN-γ production by CD56bright NK cells. J. Immunol. 196, 2552–2560 (2016).
Keppel, M. P., Saucier, N., Mah, A. Y., Vogel, T. P. & Cooper, M. A. Activation-specific metabolic requirements for NK cell IFN-γ production. J. Immunol. 194, 1954–1962 (2015).
O’Brien, K. L. & Finlay, D. K. Immunometabolism and natural killer cell responses. Nat. Rev. Immunol. 19, 282–290 (2019).
O’Sullivan, T. E., Johnson, L. R., Kang, H. H. & Sun, J. C. BNIP3- and BNIP3L-mediated mitophagy promotes the generation of natural killer cell memory. Immunity 43, 331–342 (2015).
Pearce, E. L., Poffenberger, M. C., Chang, C. H. & Jones, R. G. Fueling immunity: insights into metabolism and lymphocyte function. Science 342, 1242454 (2013).
Mills, E. L., Kelly, B. & O’Neill, L. A. J. Mitochondria are the powerhouses of immunity. Nat. Immunol. 18, 488–498 (2017).
Rambold, A. S. & Pearce, E. L. Mitochondrial dynamics at the interface of immune cell metabolism and function. Trends Immunol. 39, 6–18 (2018).
De Brito, O. M. & Scorrano, L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456, 605–610 (2008).
Cogliati, S. et al. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell 155, 160–171 (2013).
Youle, R. J. & Karbowski, M. Mitochondrial fission in apoptosis. Nat. Rev. Mol. Cell Biol. 6, 657–663 (2005).
Buck, M. D., O’Sullivan, D. & Pearce, E. L. T cell metabolism drives immunity. J. Exp. Med. 212, 1345–1360 (2015).
Yu, T., Robotham, J. L. & Yoon, Y. Increased production of reactive oxygen species in hyperglycemic conditions requires dynamic change of mitochondrial morphology. Proc. Natl Acad. Sci. USA 103, 2653–2658 (2006).
Vaupel, P. & Mayer, A. Hypoxia in tumors: pathogenesis-related classification, characterization of hypoxia subtypes, and associated biological and clinical implications. Adv. Exp. Med. Biol. 812, 19–24 (2014).
Krzywinska, E. et al. Loss of HIF-1α in natural killer cells inhibits tumour growth by stimulating non-productive angiogenesis. Nat. Commun. 8, 1597 (2017).
Van der Bliek, A. M., Shen, Q. & Kawajiri, S. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb. Perspect. Biol. 5, a011072 (2013).
Taguchi, N., Ishihara, N., Jofuku, A., Oka, T. & Mihara, K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 282, 11521–11529 (2007).
Lackner, L. L. & Nunnari, J. Small molecule inhibitors of mitochondrial division: tools that translate basic biological research into medicine. Chem. Biol. 17, 578–583 (2010).
Macia, E. et al. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell 10, 839–850 (2006).
Chen, H. et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160, 189–200 (2003).
Chen, H. & Chan, D. C. Emerging functions of mammalian mitochondrial fusion and fission. Hum. Mol. Genet. 14, R283–R289 (2005).
Schofield, C. J. & Ratcliffe, P. J. Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol. 5, 343–354 (2004).
Peng, H. et al. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J. Clin. Invest. 123, 1444–1456 (2013).
Bjorkstrom, N. K., Ljunggren, H. G. & Michaelsson, J. Emerging insights into natural killer cells in human peripheral tissues. Nat. Rev. Immunol. 16, 310–320 (2016).
Davoli, T., Uno, H., Wooten, E. C. & Elledge, S. J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 355, eaaf8399 (2017).
Youle, R. J. & van der Bliek, A. M. Mitochondrial fission, fusion, and stress. Science 337, 1062–1065 (2012).
Van der Windt, G. J. et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78 (2012).
Buck, M. D. et al. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 166, 63–76 (2016).
Cong, J. et al. Dysfunction of natural killer cells by FBP1-induced inhibition of glycolysis during lung cancer progression. Cell Metab. 28, 243–255 (2018).
Kaiser, B. K. et al. Disulphide-isomerase-enabled shedding of tumour-associated NKG2D ligands. Nature 447, 482–486 (2007).
Kopp, H. G., Placke, T. & Salih, H. R. Platelet-derived transforming growth factor-β down-regulates NKG2D thereby inhibiting natural killer cell antitumor reactivity. Cancer Res. 69, 7775–7783 (2009).
Yang, M. et al. NK cell development requires Tsc1-dependent negative regulation of IL-15-triggered mTORC1 activation. Nat. Commun. 7, 12730 (2016).
Michelet, X. et al. Metabolic reprogramming of natural killer cells in obesity limits antitumor responses. Nat. Immunol. 19, 1330–1340 (2018).
Cassidy-Stone, A. et al. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev. Cell 14, 193–204 (2008).
Heng, T. S., Painter, M. W. & Immunological Genome Project Consortium. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008).
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Davoli, T. et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155, 948–962 (2013).
Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33, 1–22 (2010).
Acknowledgements
We thank W. Tao for advice regarding RNA sequencing data analyses. This work was supported by the Natural Science Foundation of China (reference numbers 81330071, 81788101, 81872318 and 81602491) and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDPB1002 and XDA12020217).
Author information
Authors and Affiliations
Contributions
H.W., Z.T. and X.Z. conceived and conducted the project. H.W. supervised the project. X.Z. and H.W. wrote the paper. X.Z. performed the experiments and data analysis. D.J. contributed to the cell culture and mouse models. Y.Q., P.C., Y.S. and Y.J. collected tissue samples and information from patients. R.S. and B.F. contributed to the imaging analysis and interpreted the data. H.Z. performed the hypoxic experiments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Zoltan Fehervari was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–6 and Supplementary Tables 1–5.
Supplementary Video 1
Mitochondrial dynamics of live hypoxic NK cells from donor 54 during 120 min of filming.
Supplementary Video 2
Mitochondrial dynamics of live hypoxic NK cells from donor 55 during 120 min of filming.
Supplementary Video 3
Mitochondrial dynamics of live hypoxic NK cells from donor 56 during 120 min of filming.
Supplementary Video 4
Mitochondrial dynamics of live normal NK cells from donor 54 during 120 min of filming.
Supplementary Video 5
Mitochondrial dynamics of live normal NK cells from donor 55 during 120 min of filming.
Supplementary Video 6
Mitochondrial dynamics of live normal NK cells from donor 56 during 120 min of filming.
Source data
Source Data Fig. 1
Unprocessed versions of the western blots shown in Fig, 3
Source Data Fig. 2
Unprocessed versions of the western blots shown in Supplementary Fig. 3
Source Data Fig. 3
Unprocessed versions of the western blots shown in Supplementary Fig. 4
Rights and permissions
About this article
Cite this article
Zheng, X., Qian, Y., Fu, B. et al. Mitochondrial fragmentation limits NK cell-based tumor immunosurveillance. Nat Immunol 20, 1656–1667 (2019). https://doi.org/10.1038/s41590-019-0511-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-019-0511-1
This article is cited by
-
Metabolic reprogramming in the tumor microenvironment of liver cancer
Journal of Hematology & Oncology (2024)
-
Natural killer cells drive 4-1BBL positive uveal melanoma towards EMT and metastatic disease
Journal of Experimental & Clinical Cancer Research (2024)
-
NK cells as powerful therapeutic tool in cancer immunotherapy
Cellular Oncology (2024)
-
TREM-1 triggers necroptosis of macrophages through mTOR-dependent mitochondrial fission during acute lung injury
Journal of Translational Medicine (2023)
-
PINK1 protects against dendritic cell dysfunction during sepsis through the regulation of mitochondrial quality control
Molecular Medicine (2023)