Abstract
The establishment of a diverse B cell antigen receptor (BCR) repertoire by V(D)J recombination also generates autoreactive B cells. Anergy is one tolerance mechanism; it renders autoreactive B cells insensitive to stimulation by self-antigen, whereas Toll-like receptor (TLR) signaling can reactivate anergic B cells. Here, we describe a critical role of the transcription factor Ikaros in controlling BCR anergy and TLR signaling. Mice with specific deletion of Ikaros in mature B cells developed systemic autoimmunity. Ikaros regulated many anergy-associated genes, including Zfp318, which is implicated in the attenuation of BCR responsiveness by promoting immunoglobulin D expression in anergic B cells. TLR signaling was hyperactive in Ikaros-deficient B cells, which failed to upregulate feedback inhibitors of the MyD88–nuclear factor κB signaling pathway. Systemic inflammation was lost on expression of a non-self-reactive BCR or loss of MyD88 in Ikaros-deficient B cells. Thus, Ikaros acts as a guardian preventing autoimmunity by promoting BCR anergy and restraining TLR signaling.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Nutt, S. L., Hodgkin, P. D., Tarlinton, D. M. & Corcoran, L. M. The generation of antibody-secreting plasma cells. Nat. Rev. Immunol. 15, 160–171 (2015).
Wardemann, H. et al. Predominant autoantibody production by early human B cell precursors. Science 301, 1374–1377 (2003).
Zikherman, J., Parameswaran, R. & Weiss, A. Endogenous antigen tunes the responsiveness of naïve B cells but not T cells. Nature 489, 160–164 (2012).
Theofilopoulos, A. N., Kono, D. H. & Baccala, R. The multiple pathways to autoimmunity. Nat. Immunol. 18, 716–724 (2017).
Cambier, J. C., Gauld, S. B., Merrell, K. T. & Vilen, B. J. B-cell anergy: from transgenic models to naturally occurring anergic B cells? Nat. Rev. Immunol. 7, 633–643 (2007).
Rawlings, D. J., Metzler, G., Wray-Dutra, M. & Jackson, S. W. Altered B cell signalling in autoimmunity. Nat. Rev. Immunol. 17, 421–436 (2017).
Tsokos, G. C., Lo, M. S., Costa Reis, P. & Sullivan, K. E. New insights into the immunopathogenesis of systemic lupus erythematosus. Nat. Rev. Rheumatol. 12, 716–730 (2016).
Georgopoulos, K. The making of a lymphocyte: the choice among disparate cell fates and the IKAROS enigma. Genes Dev. 31, 439–450 (2017).
Han, J.-W. et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet. 41, 1234–1237 (2009).
Gateva, V. et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat. Genet. 41, 1228–1233 (2009).
He, C. F. et al. TNIP1, SLC15A4, ETS1, RasGRP3 and IKZF1 are associated with clinical features of systemic lupus erythematosus in a Chinese Han population. Lupus 19, 1181–1186 (2010).
Cunninghame Graham, D. S. et al. Association of NCF2, IKZF1, IRF8, IFIH1, and TYK2 with systemic lupus erythematosus. PLoS Genet. 7, e1002341 (2011).
Farh, K. K.-H. et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 (2015).
Zhang, Y.-M. et al. Association of the IKZF1 5′ UTR variant rs1456896 with lupus nephritis in a northern Han Chinese population. Scand. J. Rheumatol. 46, 210–214 (2017).
Kuehn, H. S. et al. Loss of B cells in patients with heterozygous mutations in IKAROS. N. Engl. J. Med. 374, 1032–1043 (2016).
Hoshino, A. et al. Abnormal hematopoiesis and autoimmunity in human subjects with germline IKZF1 mutations. J. Allergy Clin. Immunol. 140, 223–231 (2017).
Bogaert, D. J. et al. A novel IKAROS haploinsufficiency kindred with unexpectedly late and variable B-cell maturation defects. J. Allergy Clin. Immunol. 141, 432–435 (2018).
Van Nieuwenhove, E. et al. A kindred with mutant IKAROS and autoimmunity. J. Allergy Clin. Immunol. 142, 699–702 (2018).
Schwickert, T. A. et al. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat. Immunol. 15, 283–293 (2014).
Kwon, K. et al. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity 28, 751–762 (2008).
Naradikian, M. S., Hao, Y. & Cancro, M. P. Age-associated B cells: key mediators of both protective and autoreactive humoral responses. Immunol. Rev. 269, 118–129 (2016).
Rubtsova, K., Rubtsov, A. V., Cancro, M. P. & Marrack, P. Age-associated B cells: a T-bet-dependent effector with roles in protective and pathogenic immunity. J. Immunol. 195, 1933–1937 (2015).
Kallies, A., Xin, A., Belz, G. T. & Nutt, S. L. Blimp-1 transcription factor is required for the differentiation of effector CD8+ T cells and memory responses. Immunity 31, 283–295 (2009).
Gu, H., Zou, Y. R. & Rajewsky, K. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell 73, 1155–1164 (1993).
Goodnow, C. C. et al. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature 334, 676–682 (1988).
Goodnow, C. C., Crosbie, J., Jorgensen, H., Brink, R. A. & Basten, A. Induction of self-tolerance in mature peripheral B lymphocytes. Nature 342, 385–391 (1989).
Sabouri, Z. et al. IgD attenuates the IgM-induced anergy response in transitional and mature B cells. Nat. Commun. 7, 13381 (2016).
Enders, A. et al. Zinc-finger protein ZFP318 is essential for expression of IgD, the alternatively spliced Igh product made by mature B lymphocytes. Proc. Natl Acad. Sci. USA 111, 4513–4518 (2014).
Pioli, P. D., Debnath, I., Weis, J. J. & Weis, J. H. Zfp318 regulates IgD expression by abrogating transcription termination within the Ighm/Ighd locus. J. Immunol. 193, 2546–2553 (2014).
Sharma, S., Fitzgerald, K. A., Cancro, M. P. & Marshak-Rothstein, A. Nucleic acid-sensing receptors: rheostats of autoimmunity and autoinflammation. J. Immunol. 195, 3507–3512 (2015).
Nickerson, K. M. et al. TLR9 promotes tolerance by restricting survival of anergic anti-DNA B cells, yet is also required for their activation. J. Immunol. 190, 1447–1456 (2013).
Sindhava, V. J. et al. A TLR9-dependent checkpoint governs B cell responses to DNA-containing antigens. J. Clin. Invest. 127, 1651–1663 (2017).
Kobayashi, K. et al. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell 110, 191–202 (2002).
Tavares, R. M. et al. The ubiquitin modifying enzyme A20 restricts B cell survival and prevents autoimmunity. Immunity 33, 181–191 (2010).
Chu, Y. et al. B cells lacking the tumor suppressor TNFAIP3/A20 display impaired differentiation and hyperactivation and cause inflammation and autoimmunity in aged mice. Blood 117, 2227–2236 (2011).
Abdul-Sater, A. A. et al. The signaling adaptor TRAF1 negatively regulates Toll-like receptor signaling and this underlies its role in rheumatic disease. Nat. Immunol. 18, 26–35 (2017).
Schenten, D. et al. Signaling through the adaptor molecule MyD88 in CD4+ T cells is required to overcome suppression by regulatory T cells. Immunity 40, 78–90 (2014).
Shin, H. et al. A role for the transcriptional repressor Blimp-1 in CD8+ T cell exhaustion during chronic viral infection. Immunity 31, 309–320 (2009).
Hale, J. S., Boursalian, T. E., Turk, G. L. & Fink, P. J. Thymic output in aged mice. Proc. Natl Acad. Sci. USA 103, 8447–8452 (2006).
Chan, O. T., Hannum, L. G., Haberman, A. M., Madaio, M. P. & Shlomchik, M. J. A novel mouse with B cells but lacking serum antibody reveals an antibody-independent role for B cells in murine lupus. J. Exp. Med. 189, 1639–1648 (1999).
Amano, H. et al. The Yaa mutation promoting murine lupus causes defective development of marginal zone B cells. J. Immunol. 170, 2293–2301 (2003).
Degn, S. E. et al. Clonal evolution of autoreactive germinal centers. Cell 170, 913–926 (2017).
Noviski, M. et al. IgM and IgD B cell receptors differentially respond to endogenous antigens and control B cell fate. eLife 7, e35074 (2018).
Übelhart, R. et al. Responsiveness of B cells is regulated by the hinge region of IgD. Nat. Immunol. 16, 534–543 (2015).
Maity, P. C. et al. B cell antigen receptors of the IgM and IgD classes are clustered in different protein islands that are altered during B cell activation. Sci. Signal. 8, ra93 (2015).
Noviski, M. & Zikherman, J. Control of autoreactive B cells by IgM and IgD B cell receptors: maintaining a fine balance. Curr. Opin. Immunol. 55, 67–74 (2018).
Sun, H., Lu, B., Li, R.-Q., Flavell, R. A. & Taneja, R. Defective T cell activation and autoimmune disorder in Stra13-deficient mice. Nat. Immunol. 2, 1040–1047 (2001).
Li, S. et al. The transcription factors Egr2 and Egr3 are essential for the control of inflammation and antigen-induced proliferation of B and T cells. Immunity 37, 685–696 (2012).
Lech, M. et al. Interleukin-1 receptor-associated kinase-M suppresses systemic lupus erythematosus. Ann. Rheum. Dis. 70, 2207–2217 (2011).
Shlomchik, M. J., Madaio, M. P., Ni, D., Trounstein, M. & Huszar, D. The role of B cells in lpr/lpr-induced autoimmunity. J. Exp. Med. 180, 1295–1306 (1994).
Wang, J.-H. et al. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity 5, 537–549 (1996).
Kallies, A. et al. Plasma cell ontogeny defined by quantitative changes in Blimp-1 expression. J. Exp. Med. 200, 967–977 (2004).
Ohinata, Y. et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436, 207–213 (2005).
Bouvier, G. et al. Deletion of the mouse T-cell receptor β gene enhancer blocks αβ T-cell development. Proc. Natl Acad. Sci. USA 93, 7877–7881 (1996).
Shinkai, Y. et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 68, 855–867 (1992).
Driegen, S. et al. A generic tool for biotinylation of tagged proteins in transgenic mice. Transgenic Res. 14, 477–482 (2005).
Barnden, M. J., Allison, J., Heath, W. R. & Carbone, F. R. Defective TCR expression in transgenic mice constructed using cDNA-based α- and β-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 76, 34–40 (1998).
Kohwi-Shigematsu, T. et al. SATB1-mediated functional packaging of chromatin into loops. Methods 58, 243–254 (2012).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Parkhomchuk, D. et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 37, e123 (2009).
Wöhner, M. et al. Molecular functions of the transcription factors E2A and E2-2 in controlling germinal center B cell and plasma cell development. J. Exp. Med. 213, 1201–1221 (2016).
Yates, A. et al. Ensembl 2016. Nucleic Acids Res. 44, D710–D716 (2016).
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Kuhn, R. M., Haussler, D. & Kent, W. J. The UCSC genome browser and associated tools. Brief Bioinform. 14, 144–161 (2013).
Ramírez, F., Dündar, F., Diehl, S., Grüning, B. A. & Manke, T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Revilla-i-Domingo, R. et al. The B-cell identity factor Pax5 regulates distinct transcriptional programmes in early and late B lymphopoiesis. EMBO J. 31, 3130–3146 (2012).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Wagner, G. P., Kin, K. & Lynch, V. J. Measurement of mRNA abundance using RNA-Seq data: RPKM measure is inconsistent among samples. Theory Biosci. 131, 281–285 (2012).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 15, 550 (2014).
Acknowledgements
We thank R. Medzhitov, A. Tarakhovsky and S. L. Nutt for providing the Myd88fl/fl, Prdm1fl/fl and Prdm1Gfp/+ mice, respectively; J. Cyster for providing the HyHEL9 antibody; G. Schmauß and M. Weninger for fluorescence-activated cell sorting; A. Sommer’s team at the Vienna BioCenter Core Facilities for Illumina sequencing; and A. Kavirayani’s team at the Vienna BioCenter Core Facilities for histology support and pathology review. This research was supported by Boehringer Ingelheim, the European Research Council (under the European Union’s Horizon 2020 research and innovation program (grant agreement number 740349-PlasmaCellControl)), the Austrian Industrial Research Promotion Agency (Headquarter Grant FFG-852936) and the Austrian Science Fund (T838-B26 to T.A.S.).
Author information
Authors and Affiliations
Contributions
T.A.S. performed almost all of the experiments. H.T. performed the Bio-ChIP-Seq experiments. K.S. performed the Phosflow staining analyses. M.F. performed the bioinformatics analysis of all of the RNA-Seq data. M.J. performed the bioinformatics analysis of all of the ChIP-Seq data. T.A.S. and M.B. planned the project, designed the experiments and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Related to Fig. 1. Splenomegaly in mice with conditional loss of Ikaros in mature B cells.
a, Photographs of the spleen, thymus and lymph nodes of 6-week-old Ikzf1B– (Cd23-Cre Ikzf1fl/–) and Ikzf1B+ (Cd23-Cre Ikzf1fl/+) mice. b, Analysis of mixed bone marrow chimeras, which were generated by transplantation of sub-lethally irradiated Rag2–/– host mice with a 1:1 mixture of bone marrow from CD45.1+ wild-type (WT) mice and bone marrow from CD45.2+ Ikzf1B– or CD45.2+ Ikzf1B+ mice. The weight of the spleen is shown for chimeric mice of the two different genotypes. The frequencies of the MZ and CD21–CD23– B cell subsets among total B cells were determined by flow cytometry. The contribution of the CD45.1+ wild-type and CD45.2+ Ikzf1B– hematopoietic progenitors to the MZ B and CD21–CD23– B cell populations in ‘Ikzf1B–‘ chimeras is shown to the right. Lines connect the two data points obtained for the wild-type and Ikzf1B– MZ or CD21–CD23– B cells analyzed within the same chimeric mouse. The chimeric mice (n = 6 for each genotype) were analyzed 4-10 weeks post transplantation c, Statistical analysis of the frequency of B-1, FO B, MZ B and CD21–CD23– B cells as well as activated effector/memory T cells (relative to total live splenocytes) in Ikzf1B+ (gray) and Ikzf1B– (black) mice (n = 2-28) at the indicated age. The spleen weight is shown as reference. d, Expression of the indicated cell surface markers and transcription factors in FO B and CD21–CD23– B cells of Ikzf1B+ and Ikzf1B– mice at the age of 6-10 weeks, as determined by flow cytometric analysis. The expression pattern of large (FSChi) CD21–CD23– B cells is shown in red. Plasmablasts (PB), dendritic cells (DC) and B-1a cells were used as reference cell types for the Blimp-1-GFP, CD11b, CD11c and CD5 staining (thin gray line), respectively. T-bet expression was analyzed by intracellular flow cytometry, and Blimp-1-GFP expression was determined in CD21–CD23– B cells from Prdm1Gfp/+ Ikzf1B– and Prdm1Gfp/+ Ikzf1B+ mice. Data in a are representative of 5-8-week-old mice analyzed in 36 independent experiments (corresponding to Fig. 1a). Data in b and c were obtained in 3 and 23 independent experiments, respectively. Data in d are representative of 2 independent experiments. Statistical data (b,c) are shown as mean values with SEM and were analyzed by the Student’s t-test (unpaired, two-tailed; b) or by multiple t-tests (unpaired, two-tailed, Holm-Sidac; c): *P < 0.03, **P < 0.002, ***P < 0.0002, ****P < 0.0001. See Source Data for exact description of the mouse numbers n (c) and P values. Each dot or connected dots correspond to one mouse. The different cell types are defined as described in the Online Methods.
Supplementary Figure 2 Related to Fig. 2. Activation of T cells and loss of GC B cells and plasma cells in Ikzf1B– mice.
a, Flow cytometric determination of the frequencies of CD4+ and CD8+ T cells and their activated effector/memory T cell subsets (CD44+CD62L–; blue shading) among total live cells in the spleen of Ikzf1B– (black, n = 4 or 3) and Ikzf1B+ (gray, n = 4 or 3) mice at the age of 2 weeks or > 18 weeks. b, Cytokine concentrations in the serum of 4-8-week-old Ikzf1B– (black, n = 5) and Ikzf1B+ (gray, n = 7) mice, shown as fluorescence intensity (arbitrary units, AU). The following concentrations of the cytokine standards are indicated as dashed line: IFN-γ (1 pg/ml), TNF (3 pg/ml), IL-23 (13 pg/ml), IL-10 (2 pg/ml), IL-1α (25 pg/ml) and IL-6 (5 pg/ml). c, Immunohistological analysis of spleen sections from 9-week-old mice of the indicated genotypes. Staining with anti-IgM (green), anti-IgD (blue), and anti-CD169/MOMA-1 (red) antibodies highlights MZ B cells (arrowhead), FO B cells and the metallophilic macrophages (separating the MZ and FO B cell zones) in Ikzf1B+ mice, respectively. The T cell zone (T) is indicated. d, Infiltration of the liver by B220+ B and CD3+ T cells in a 10-week-old Ikzf1B– mouse in contrast to a control Ikzf1B+ littermate. Foci of infiltrating B and T cells (arrowheads) are present next to a dilated central vein (asterisk) in an Ikzf1B– liver. Sections were stained with hematoxylin & eosin (H&E) or anti-B220 and anti-CD3 antibodies, and a magnification of the indicated area is shown to the right. e, Statistical evaluation of the lymphocyte infiltration in the liver and kidney of Ikzf1B mice (n = 2) and their control Ikzf1B+ littermates (n = 2) at the age of 8 and 10 weeks. The numbers of discrete nodular CD3+ foci per 5 mm2 are shown as mean values with SEM. f, Flow cytometric analysis of GC B cells (gated on CD19+CD21lo B cells) of unimmunized Ikzf1B– and Ikzf1B+ mice at the age of 7 weeks. g, Immunohistological and flow cytometric analyses of the spleen from Ikzf1B+, Ikzf1B– and Ikzf1GCB– (Aicda-Cre Ikzf1fl/–) mice 9-14 days after NP-KLH immunization (in alum). Spleen sections (left) from 12-week-old Ikzf1B+ and Ikzf1B– mice (upper panel) were stained with anti-IgD (green, non-GC B cells), anti-CD3 (blue, T cells) and anti-CD35 (red, follicular dendritic cells), while spleen sections from 10-week-old Ikzf1B+ and Ikzf1GCB– (lower panel) were stained with anti-IgD (green, non-GC B cells), anti-B220 (blue, B cells), and PNA (red, GC B cells). Bar graphs (middle) indicate the frequency of GC B cells (GL7+CD95+CD19+) among total live cells in the spleen of immunized Ikzf1GCB– (black, n = 15) and Ikzf1B– (n = 7) mice at the age of 10-30 weeks. Immunized Aicda-Cre Ikzf1fl/+ mice were additionally included in the ‘Ikzf1B+’ mice (gray, n = 44). The histograms (right) display the intracellular Ikaros levels detected in the indicated cell types of Ikzf1B– (black line), Aicda-Cre Ikzf1fl/– (dashed line) and control Ikzf1fl/+ (gray shaded) mice at the age of 10-15 weeks. h,i, Flow cytometric analysis of CD138+Blimp-1+ plasma cells (PC) in the bone marrow (h) and CD138+Blimp-1+ plasmablasts (PB) in the spleen (i) of Prdm1Gfp/+ Ikzf1B– and Prdm1Gfp/+ Ikzf1B+ mice at the age of 5 weeks (upper row) and one year (lower row). Histograms (i, right) display intracellular Ikaros levels in splenic plasmablasts of the indicated genotypes. j, Titers of the indicated antibody isotypes in the serum of 4-8-week-old Ikzf1B– (black, n = 12) and Ikzf1B+ (gray, n = 15) mice, which were determined as described in the Online Methods. The data are shown as box plots with minimal to maximal whiskers including all data points. Data in a, e and g (bar graph) were generated in 3, 2 and 4 independent experiments, respectively. Data in b and j were obtained by collecting sera in 2 and 4 independent experiments, respectively, which were then analyzed in one multiplex experiment each. Data in c, f, g, h and i are representative of 3, 5, 3, 3, and 8 independent experiments, respectively. Statistical data in (b,g,j) are shown as mean values with SEM, and were analyzed by the Student’s t-test (unpaired, two-tailed; b), by one-way ANOVA with(Tukey; (g) or by multiple t-tests (unpaired, two-tailed, Holm-Sidac; j): *P < 0.03, **P < 0.002, ****P < 0.0001. Each dot corresponds to one mouse (a,b,g,j) or repeated measurements (e).
Supplementary Figure 3 Related to Fig. 3. Polyclonal T cells, but not plasma cells, induce inflammation in Ikzf1B– mice.
a, Determination of the spleen weight and relative abundance of splenic plasmablasts (PB), effector/memory T, FO B and CD21lo/–CD23– B cells in experimental Ikzf1B– Prdm1B– mice (dark green, n = 8-10) and control Ikzf1B+ Prdm1B+ (gray, n = 7-8), Ikzf1B– Prdm1B+ (black, n = 12-14) and Ikzf1B+ Prdm1B– (light green, n = 7-9) mice at the age of 5-9 weeks. The frequency of the different cell types among total live splenocytes was determined by flow cytometry. b, Flow cytometric analysis of FO and CD21lo/–CD23– B cells (gated on B-2 cells), which were quantified in a. c, Relative frequency of splenic αβ T, NK, FO B and CD21lo/–CD23– B cells in experimental Eβ–/– Ikzf1B–mice (dark blue, n = 4-6) and control Eβ+/– Ikzf1B+ (gray, n = 5-8), Eβ+/– Ikzf1B– (black, n = 3-5) and Eβ–/– Ikzf1B+ (light blue, n = 10-13) mice at the age of 5-6 weeks. The frequency of the different cell types among total live splenocytes was determined by flow cytometry. d, Flow cytometric analysis of MZ B, CD21lo/–CD23– B, CD4+ T and CD8+ T cells from the spleen of JHT mice that were reconstituted with bone marrow from Ikzf1B+ mice (gray, n = 5 (untreated), 4 (anti-CD4), 3 (anti-CD8)) or Ikzf1B– mice (black, n = 4 (untreated), 4 (anti-CD4), 4 (anti-CD8)), followed by intravenous injection of anti-CD4 or anti-CD8 antibodies at regular intervals (after day 1, week 1, 2, 3 and 4). The frequency of the different B and T cell types was determined 4.5 or 5 weeks after transplantation. e, Frequency of the indicated B and T cell types in the spleen of chimeric mice, which were generated by transplantation of Eβ–/– Ikzf1B– (dark blue) or Eβ–/– Ikzf1B+ (light blue) donor bone marrow into sub-lethally irradiated JHT (n = 4-12), Rag2–/– (n = 6-16) or OT-II TCR-tg Rag2–/– (n = 2-5) host mice. The bone marrow chimeras were analyzed by flow cytometry 4-5 weeks post transplantation. Data in b are representative of mice analyzed in a, which were examined in 7 independent experiments. Data in c, d and e were analyzed in 5, 2 and 3 independent experiments, respectively. Statistical data (a,c,d,e) are shown as mean value with SEM and were analyzed by one-way ANOVA (Tukey; a,c,e), by multiple t-test (unpaired, two-tailed, Holm-Sidak; d) or by the Student’s t-test (unpaired, two-tailed; e (Eff/Mem T)); *P < 0.03, **P < 0.002, ***P < 0.0002, ****P < 0.0001. See Source Data for exact description of the mouse numbers (n) and P values. Each dot corresponds to one mouse.
Supplementary Figure 4 Related to Fig. 4. Expression of the monoclonal MD4 BCR prevents inflammation in Ikzf1B– mice.
a, Antigen presentation by Ikaros-deficient FO B cells. Splenic CD23+ FO B cells from control Ikzf1B+ mice as well as CD23+ (FO B) and CD21lo/–CD23– B cells from inflamed Ikzf1B– mice at the age of 8 weeks were loaded with the OTII peptide before similar numbers of live cells of each B cell type were co-cultured for 4 days at a 1:1 ratio with naïve splenic CD4+ OTII TCR-tg Rag2–/– T cells that were labeled with CellTrace Violet (see Online Methods). The proliferation of T cells (CellTrace Violet dilution) is shown to the left, while the differentiation of naïve T cells (CD44–CD62L+) to activated T cells (CD44+CD62L–) under the control of the Ikaros-deficient CD23+ FO or CD21low/–CD23– B cells is shown to the right (gated on live Vβ5.1/2+ Viability Dye eF780– cells). b, Immunohistological analysis of spleen sections from 6-week-old MD4 BCR-tg Ikzf1B+ and MD4 BCR-tg Ikzf1B– littermates, which were stained with anti-TCRβ (blue, T cells), anti-IgD (green, B cells) and anti-CD169/MOMA-1 (red, metallophilic macrophages) antibodies. c, Expression of the indicated cell surface proteins on FO B cells (CD19+CD5–CD93–CD21loCD23+) and MZ B cells (CD19+CD5–CD93–CD21+CD23lo) of MD4 BCR-tg Ikzf1B+ (purple surface) or MD4 BCR-tg Ikzf1B– (purple line) littermates, as determined by flow cytometry. For analysis of CD21 (Cr2) expression, FO and MZ B cells were gated as CD19+CD5–CD93–CD23+CD1dlo and CD19+CD5–CD93–CD23loCD1d+, respectively. T1 B cells of the MD4 BCR-tg Ikzf1B+ genotype are shown for comparison. Data in a, b and c are representative of 2, 2 and 3 independent experiments, respectively.
Supplementary Figure 5 Related to Fig. 5. Ikaros controls anergy induction upon chronic antigen exposure.
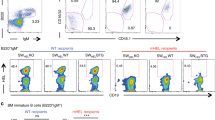
a, Intracellular BCR signaling. The phosphorylation (p-) status of signal transducers downstream of the BCR was determined in lymph node FO B cells of MD4 BCR-tg Ikzf1B+ (purple shaded or dashed line) and MD4 BCR-tg Ikzf1B– (purple line) before (unstimulated (unst.), dashed line) and after 5 min of anti-IgM addition or 15 min of HEL stimulation. Flow cytometric analysis was performed with antibodies specific for p-SYK (p-Tyr525/526), p-BLNK (p-Tyr84), p-PLCγ2 (p-Tyr759), p-ERK1/2 (p-Thr202/204), p-AKT (p-Ser473) and p-S6 (p-Ser235/236). b, Measurement of intracellular Ca2+ fluxes in HEL-stimulated CD43– FO B cells from lymph nodes of MD4 BCR-tg Ikzf1B+ or MD4 BCR-tg Ikzf1B–mice, which were either not pre-treated (naïve), were injected with HEL antigen for 1 day or additionally carried the ML5 transgene. c, Evaluation of the Ikzf1 deletion frequency by intracellular Ikaros staining of transitional T1 B cells and mature B cells (CD19+CD5–CD138–CD93–CD23+IgD+) from the spleen of MD4 BCR-tg Ikzf1B+ (purple shaded) and MD4 BCR-tg Ikzf1B– (purple line) mice lacking (upper row) or containing the ML5 transgene (lower row). d, HEL binding to the MD4 BCR receptor at 1, 4 and 20 hours after HEL injection into MD4 BCR-tg Ikzf1B– and control MD4 BCR-tg Ikzf1B+ mice, as shown by flow cytometric analysis of lymph node B cells (CD19+CD138–) with an anti-HEL antibody (HyHEL9). Data in a, c and d are representative for 3, 4 and 2 independent experiments, respectively. Data in b are representative of experiments analyzed in Fig. 5a, c, e.
Supplementary Figure 6 Related to Fig. 6. Ikaros-regulated genes in anergic B cells.
a, Flow cytometric sorting of splenic FO B cells as exemplified for MD4 BCR-tg ML5-tg Ikzf1B+ FO B cells. After immunomagnetic depletion with CD43-MicroBeads, FO B cells were sorted as CD19+CD23+CD21int CD93int/–TCRβ–CD138– cells. Reanalysis determined a purity of > 95% (here 98.5%) for the sorted live FO B cells. b, Absence of exon 8 in Ikzf1 transcripts following gene inactivation. The combined RNA-seq profile of all replicate experiments is shown for naïve and 1-day HEL-exposed FO B cells of the indicated genotypes. The floxed exon 8 is marked by gray shading. c,d, Scatter plot of gene expression differences between naïve FO B cells of the control MD4 BCR-tg Ikzf1B+ and Ikaros mutant MD4 BCR-tg Ikzf1B– genotypes (c) as well as between 1-day HEL-exposed FO B cells of same control and Ikaros mutant genotypes (d). The expression of each gene was plotted as normalized log10 (norm rlog) expression value (see Online Methods). Ikaros-regulated genes were defined by an expression difference of > 2-fold, an adjusted P value of < 0.05 and a TPM (transcripts per million) value of > 5 in B cells of the control or Ikaros mutant genotype, respectively, which resulted in the identification of the indicated number of Ikaros-activated (blue) and Ikaros-repressed (red) genes in naïve FO B cells (c; Table 1) or HEL-exposed FO B cells (d; Table 2). The dot size varies according to the adjusted P value calculated for each gene. e, Overlap of the Ikaros-dependent gene expression patterns between naïve and 1-day HEL-exposed FO B cells (Table 3). The log2-fold expression change observed between HEL-exposed Ikaros-deficient (MD4 BCR-tg Ikzf1B–) and control (MD4 BCR-tg Ikzf1B+) FO B cells (horizontal axis) as well as between naive Ikaros-deficient and control FO B cells (vertical axis) is plotted for each gene. Differentially expressed genes, which passed the above mentioned selection criteria (c,d), are marked by dark blue dots (Ikaros-activated) or dark red dots (Ikaros-repressed) in the naïve FO B cell comparison or by blue open circles (Ikaros-activated) and red open circles (Ikaros-repressed) in the comparison of the HEL-exposed FO B cells. The size of the circles varies according to the adjusted P value calculated for each gene in HEL-exposed FO B cells. A Pearson coefficient of 0.67 and a regression slope of 0.53 were calculated for the comparison of the Ikaros-dependent gene expression patterns between naïve and HEL-induced FO B cells. Based on the above mentioned selection criteria (c,d), 42 Ikaros-repressed and 15 Ikaros-activated genes overlapped in naïve and HEL-treated FO B cells (closed circles). f, Functional classification and quantification of the proteins that are encoded by Ikaros-activated and Ikaros-repressed genes of the Ikaros-dependent gene expression signature (Fig. 6a, Table 3). The bar size indicates the percentage of activated or repressed genes in each functional class relative to the total activated or repressed genes, respectively. Numbers in the bars indicate the number of genes in each functional class. g, Definition of an RNA-seq-based anergy expression signature (volcano plot, lower part) and its correlation with a previously published anergy expression signature (density profile, upper part). Based on RNA-seq analyses of naïve MD4 BCR-tg FO B cells and fully anergic MD4 BCR-tg ML5-tg FO B cells, an anergy expression signature was defined by selecting genes with an expression difference of > 3-fold, an adjusted P value of < 0.05 and a TPM value of > 5 in naïve or anergic FO B cells, respectively, which identified 59 genes (green dots) with increased expression (‘anergy up’) and 50 genes (brown dots) with decreased expression (‘anergy down’) in anergic FO B cells compared to naïve B cells (Table 4). Black circles denote genes, which are described as ‘strongly’ regulated genes in the anergy expression signature of Sabouri et al. (Nat. Commun. 7, 13381). A density profile (upper part) shows the correlation of the published ‘strongly’ and ‘moderately-to-strongly’ regulated anergy signature genes with the log2-fold expression changes of the differentially expressed genes defined by the RNA-seq comparison of naïve and anergic FO B cells. h, Gene set enrichment analysis (GSEA) of the 59 ‘anergy up’ genes (upper panel) and 50 ‘anergy down’ genes (lower panel) based on the ranked log2-fold gene expression changes determined for Ikaros-deficient versus control HEL-exposed FO B cells. NES, normalized enrichment score; FDR, false discovery rate. i, Expression of Ighm and Ighd mRNAs (coding for IgM and IgD) in naïve or HEL-exposed Ikaros-deficient FO B cells (dark purple) and in naïve or HEL-exposed control FO B cells (light purple). Gray bars display Ighm and Ighd expression in fully anergic MD4 BCR-tg ML5-tg FO B cells, while the light purple bar overlaid on the gray bar indicates the corresponding expression in naïve MD4 BCR-tg FO B cells. Data in a are representative of the sorting of FO B cells, used for RNA-seq analysis. The RNA-seq data shown in b-i were obtained from 2 (naïve FO B) and 5 (HEL-exposed FO B) independent experiments. Statistical data (i) are shown as mean values with SEM.
Supplementary Figure 7 Related to Fig. 7. Hyperactivation of Ikaros-deficient B cells upon TLR stimulation.
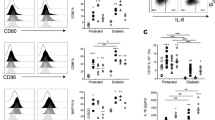
a, Expression of the different Tlr genes in naïve FO B cells of the indicated genotypes, as determined by RNA-seq. b, Time course analysis of B cell activation in response to LPS, R848 or CpG stimulation. CD43– FO B cells isolated from the spleen of Ikaros mutant (MD4 BCR-tg Ikzf1B–; dark purple line) and control (MD4 BCR-tg Ikzf1B+; light purple) mice were stained with the CellTrace Violet reagent prior to LPS, R848 or CpG stimulation for up to 4 days. The CellTrace Violet staining, cell size and expression of the activation marker CD69 were analyzed at the indicated time points by flow cytometry. Unstimulated ex vivo sorted FO B cells are shown as a reference. c, Schematic diagram of the stimulatory CpG-HEL immune complex (SCHIC), which was generated by mixing HEL-Bio1-2, CpG-Bio and streptavidin (SA) at a 1:1:2 molar ratio (see Online Methods). d, Survival of CD43– FO B cells of the indicated genotypes after stimulation with a HEL-Bio-SA complex (n = 10), CpG-Bio (n = 10) and the SCHIC reagent (n = 5) for 3 or 4 days, as determined by staining with the viability dye eF780. e, Activation of the CD43– FO B cells of the indicated genotypes with the SCHIC reagent for up to 4 days. The proliferation and survival of the FO B cells were measured by staining with CellTrace Violet and the viability dye eF780, respectively (upper panel). The viable FO B cells were further analyzed for their cell size and CD69 expression by flow cytometry (lower panel). Dead cells are indicated by blue shading. The RNA-seq data presented in a were obtained from 2 independent experiments and are shown as mean values with SEM. The data in b, d and e are representative of 3, 2 and 2 independent experiments. Statistical data in d are shown as box plots with minimal to maximal whiskers including all data points and were analyzed by multiple t-test (unpaired, two-tailed, Holm-Sidak); *P < 0.03.
Supplementary Figure 8 Related to Fig. 7. Ikaros-regulated genes in CpG-stimulated FO B cells.
a, Heat map displaying the expression of Ikaros-dependent genes in 1-day CpG-stimulated FO B cells, which were identified as significantly regulated between the Ikaros mutant (MD4 BCR-tg Ikzf1B–) and control (MD4 BCR-tg Ikzf1B+) B cells (Fig. 7b). Expression of the same genes is additionally shown for naïve and HEL-exposed FO B cells of the Ikaros mutant or control genotype. Genes that were regulated significantly in the same direction as in naïve and/or HEL-exposed FO B cells were grouped into ‘common regulation’, while genes with no significant or opposite regulation in naïve and/or HEL-exposed FO B cells were grouped into ‘unique regulation’. The ‘unique regulation’ group was subdivided into genes that were not expressed (TPM < 5) or expressed (TPM > 5) in naïve and HEL-exposed FO B cells. For detailed information about the grouping see Table 5 (‘group code’). The gene expression values (TPM) in Ikaros-deficient and control FO B cells are displayed according to the color keys shown to the right (top). Within groups, genes were sorted according to their expression difference (ΔTPM) in CpG-stimulated B cells. Black squares denote Ikaros binding at the indicated genes in naïve FO B cells. b, Functional classification and quantification of the proteins that are encoded by Ikaros-activated and Ikaros-repressed genes in 1-day CpG-stimulated FO B cells (Fig. 7b, Table 5). The bar size indicates the percentage of activated or repressed genes in each functional class relative to the total number of activated or repressed genes, respectively. Numbers in the bars indicate the number of genes in each functional class. c, Reduced expression of the TNFAIP3 protein in Ikaros-deficient FO B cells upon stimulation with CpG or R848 for the indicated time. The expression levels of the TNFAIP3 and reference GAPDH proteins were determined by immunoblot analysis of stimulated MD4 BCR-tg Ikzf1B– and control MD4 BCR-tg Ikzf1B+ B cells. The size of marker proteins is indicated in kilodaltons to the left. d, Expression of the Ikaros-activated gene Irak3 and the Ikaros-repressed genes Aif1, Tmem176b, Wnt10a, Serpinf, and Rassf4 in naïve and 1-day CpG-stimulated FO B cells of the control (MD4 BCR-tg Ikzf1B+, light purple) or Ikaros mutant (MD4 BCR-tg Ikzf1B–, dark purple) genotype, as measured by RNA-seq (upper part). Ikaros binding (black) was determined by Bio-ChIP-seq analysis of naïve Ikzf1ihCd2/ihCd2 Rosa26BirA/ BirA FO B cells, and open chromatin regions were mapped by ATAC-seq of control (MD4 BCR-tg Ikzf1B+, light purple, n = 2) and Ikaros-deficient (MD4 BCR-tg Ikzf1B–, dark purple, n = 2) FO B cells after 1 day of CpG stimulation (lower part). The Ikaros peaks were identified by ‘peak calling’ (horizontal bars). Yellow shading highlights the appearance of new open chromatin regions in CpG-stimulated Ikaros-deficient FO B cells at sites, where Ikaros was bound in control FO B cells. e, Flow cytometric analysis of mature splenic B-2 cell subsets (CD19+CD5–CD138–CD93–) in polyclonal Ikzf1B– (Cd23-Cre Myd88+/+ Ikzf1fl/fl) and Myd88B–/– Ikzf1B– (Cd23-Cre Myd88fl/fl Ikzf1fl/fl) mice. The percentage of cells in each gate is indicated. The RNA-seq data shown in a,b,d were obtained from 2 (naïve FO B) and 4 (CpG-stimulated FO B) independent experiments. The ChIP-seq data in d represent the average values of 4 independent experiments. Data in c are representative of 2 independent experiments, and data in e are representative of 7 independent experiment analyzed in Fig. 7f. Statistical data are shown as mean values with SEM (d).
Supplementary information
Supplementary Table 1
Ikaros-regulated genes determined by RNA-Seq of ex vivo-sorted Ikzf1B+ (control) and Ikzf1B− (Ikaros-knockout) naive FO B cells.
Supplementary Table 2
Ikaros-regulated genes determined by RNA-Seq of ex vivo-sorted Ikzf1B+ (control) and Ikzf1B– (Ikaros-knockout) HEL-exposed (1 d; in vivo) FO B cells.
Supplementary Table 3
Gene annotation and description of Ikaros-regulated genes in ex vivo-sorted naive and HEL-exposed FO B cells.
Supplementary Table 4
Anergy signature genes determined by RNA-Seq of naive MD4 BCR-tg (wild-type MD4) and anergic MD4 BCR-tg ML5-tg (wild-type MD4 ML5) B cells.
Supplementary Table 5
Ikaros-regulated genes determined by RNA-Seq of sorted Ikzf1B+ (control) and Ikzf1B– (Ikaros-knockout) CpG-stimulated (1 d; in vitro) FO B cells.
Supplementary Table 6
Description of all Illumina sequencing experiments generated for this study.
Rights and permissions
About this article
Cite this article
Schwickert, T.A., Tagoh, H., Schindler, K. et al. Ikaros prevents autoimmunity by controlling anergy and Toll-like receptor signaling in B cells. Nat Immunol 20, 1517–1529 (2019). https://doi.org/10.1038/s41590-019-0490-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-019-0490-2
This article is cited by
-
Inhibition of STAT3 alleviates LPS-induced apoptosis and inflammation in renal tubular epithelial cells by transcriptionally down-regulating TASL
European Journal of Medical Research (2024)
-
Death-associated protein 3 in cell death and beyond
Genome Instability & Disease (2024)
-
A DEAD-box helicase drives the partitioning of a pro-differentiation NAB protein into nuclear foci
Nature Communications (2023)
-
TLR7 gain-of-function genetic variation causes human lupus
Nature (2022)
-
Management of Chronic Immune Thrombocytopenia and Presumed Autoimmune Hepatitis in a Child with IKAROS Haploinsufficiency
Journal of Clinical Immunology (2020)